Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation
- PMID: 22590557
- PMCID: PMC3349715
- DOI: 10.1371/journal.pone.0036530
Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation
Abstract
Background: Lung adenocarcinoma (LAD) has extreme genetic variation among patients, which is currently not well understood, limiting progress in therapy development and research. LAD intrinsic molecular subtypes are a validated stratification of naturally-occurring gene expression patterns and encompass different functional pathways and patient outcomes. Patients may have incurred different mutations and alterations that led to the different subtypes. We hypothesized that the LAD molecular subtypes co-occur with distinct mutations and alterations in patient tumors.
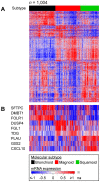
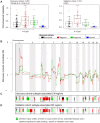
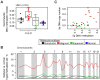
Methodology/principal findings: The LAD molecular subtypes (Bronchioid, Magnoid, and Squamoid) were tested for association with gene mutations and DNA copy number alterations using statistical methods and published cohorts (n = 504). A novel validation (n = 116) cohort was assayed and interrogated to confirm subtype-alteration associations. Gene mutation rates (EGFR, KRAS, STK11, TP53), chromosomal instability, regional copy number, and genomewide DNA methylation were significantly different among tumors of the molecular subtypes. Secondary analyses compared subtypes by integrated alterations and patient outcomes. Tumors having integrated alterations in the same gene associated with the subtypes, e.g. mutation, deletion and underexpression of STK11 with Magnoid, and mutation, amplification, and overexpression of EGFR with Bronchioid. The subtypes also associated with tumors having concurrent mutant genes, such as KRAS-STK11 with Magnoid. Patient overall survival, cisplatin plus vinorelbine therapy response and predicted gefitinib sensitivity were significantly different among the subtypes.
Conclusions/ significance: The lung adenocarcinoma intrinsic molecular subtypes co-occur with grossly distinct genomic alterations and with patient therapy response. These results advance the understanding of lung adenocarcinoma etiology and nominate patient subgroups for future evaluation of treatment response.
Conflict of interest statement
Figures



Similar articles
-
Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities.Cancer Discov. 2015 Aug;5(8):860-77. doi: 10.1158/2159-8290.CD-14-1236. Epub 2015 Jun 11. Cancer Discov. 2015. PMID: 26069186 Free PMC article.
-
Identification of transcriptional subgroups in EGFR-mutated and EGFR/KRAS wild-type lung adenocarcinoma reveals gene signatures associated with patient outcome.Clin Cancer Res. 2013 Sep 15;19(18):5116-26. doi: 10.1158/1078-0432.CCR-13-0928. Epub 2013 Aug 12. Clin Cancer Res. 2013. PMID: 23938291
-
Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study.PLoS Med. 2016 Dec 6;13(12):e1002162. doi: 10.1371/journal.pmed.1002162. eCollection 2016 Dec. PLoS Med. 2016. PMID: 27923066 Free PMC article.
-
Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts.J Clin Oncol. 2006 Nov 1;24(31):5079-90. doi: 10.1200/JCO.2005.05.1748. J Clin Oncol. 2006. PMID: 17075127 Review.
-
Throwing new light on lung cancer pathogenesis: updates on three recent topics.Cancer Sci. 2005 Feb;96(2):63-8. doi: 10.1111/j.1349-7006.2005.00021.x. Cancer Sci. 2005. PMID: 15723649 Free PMC article. Review.
Cited by
-
DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2.Oncogene. 2015 Nov 26;34(48):5869-78. doi: 10.1038/onc.2015.38. Epub 2015 Mar 9. Oncogene. 2015. PMID: 25746006 Free PMC article.
-
Lung adenocarcinoma in the era of targeted therapies: histological classification, sample prioritization, and predictive biomarkers.Clin Transl Oncol. 2013 Jul;15(7):503-8. doi: 10.1007/s12094-012-0983-z. Epub 2013 Jan 29. Clin Transl Oncol. 2013. PMID: 23359174 Free PMC article.
-
Tumor microenvironment-associated lactate metabolism regulates the prognosis and precise checkpoint immunotherapy outcomes of patients with lung adenocarcinoma.Eur J Med Res. 2022 Nov 21;27(1):256. doi: 10.1186/s40001-022-00895-6. Eur J Med Res. 2022. PMID: 36411477 Free PMC article.
-
Lung Cancer Gene Signatures and Clinical Perspectives.Microarrays (Basel). 2013 Dec 13;2(4):318-39. doi: 10.3390/microarrays2040318. Microarrays (Basel). 2013. PMID: 27605195 Free PMC article. Review.
-
Identification of microRNAs involved in pathways which characterize the expression subtypes of NSCLC.Mol Oncol. 2019 Dec;13(12):2604-2615. doi: 10.1002/1878-0261.12571. Epub 2019 Sep 22. Mol Oncol. 2019. PMID: 31505091 Free PMC article.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

