Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation
- PMID: 22582061
- PMCID: PMC3416363
- DOI: 10.1128/AEM.00588-12
Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation
Abstract
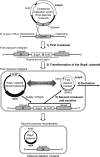
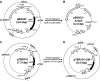
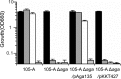
Functional analysis of Bifidobacterium genes is essential for understanding host-Bifidobacterium interactions with beneficial effects on human health; however, the lack of an effective targeted gene inactivation system in bifidobacteria has prevented the development of functional genomics in this bacterium. Here, we report the development of a markerless gene deletion system involving a double crossover in Bifidobacterium longum. Incompatible plasmid vectors were used to facilitate a second crossover step. The conditional replication vector pBS423-ΔrepA, which lacks the plasmid replication gene repA, was integrated into the target gene by a first crossover event. Subsequently, the replicative plasmid pTBR101-CM, which harbors repA, was introduced into this integrant to facilitate the second crossover step and subsequent elimination of the excised conditional replication vector from the cells by plasmid incompatibility. The proposed system was confirmed to work as expected in B. longum 105-A using the chromosomal full-length β-galactosidase gene as a target. Markerless gene deletion was tested using the aga gene, which encodes α-galactosidase, whose substrates include raffinose. Almost all the pTBR101-CM-transformed strains became double-crossover recombinants after subculture, and 4 out of the 270 double-crossover recombinants had lost the ability to assimilate raffinose. Genotype analysis of these strains revealed markerless gene deletion of aga. Carbohydrate assimilation analysis and α-galactosidase activity measurement were conducted using both the representative mutant and a plasmid-based aga-complemented strain. These functional analyses revealed that aga is the only gene encoding a functional α-galactosidase enzyme in B. longum 105-A.
Figures

Similar articles
-
A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum.Appl Microbiol Biotechnol. 2012 Jul;95(2):499-509. doi: 10.1007/s00253-012-4090-4. Epub 2012 May 27. Appl Microbiol Biotechnol. 2012. PMID: 22639142
-
Functional analysis of bifidobacterial promoters in Bifidobacterium longum and Escherichia coli using the α-galactosidase gene as a reporter.J Biosci Bioeng. 2014 Nov;118(5):489-95. doi: 10.1016/j.jbiosc.2014.05.002. Epub 2014 Jun 3. J Biosci Bioeng. 2014. PMID: 24932968
-
Synthesis of Stachyobifiose Using Bifidobacterial α-Galactosidase Purified from Recombinant Escherichia coli.J Agric Food Chem. 2018 Feb 7;66(5):1184-1190. doi: 10.1021/acs.jafc.7b04703. Epub 2018 Jan 24. J Agric Food Chem. 2018. PMID: 29363955
-
Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector.Appl Environ Microbiol. 2006 Jan;72(1):527-35. doi: 10.1128/AEM.72.1.527-535.2006. Appl Environ Microbiol. 2006. PMID: 16391088 Free PMC article.
-
Technological advances in bifidobacterial molecular genetics: application to functional genomics and medical treatments.Biosci Microbiota Food Health. 2012;31(2):15-25. doi: 10.12938/bmfh.31.15. Epub 2012 Apr 20. Biosci Microbiota Food Health. 2012. PMID: 24936345 Free PMC article. Review.
Cited by
-
The importance of genetic research on the dominant species of human intestinal indigenous microbiota.Biosci Microbiota Food Health. 2021;40(1):19-26. doi: 10.12938/bmfh.2020-011. Epub 2020 Sep 10. Biosci Microbiota Food Health. 2021. PMID: 33520565 Free PMC article. Review.
-
A Resource for Cloning and Expression Vectors Designed for Bifidobacteria: Overview of Available Tools and Biotechnological Applications.Methods Mol Biol. 2021;2278:157-182. doi: 10.1007/978-1-0716-1274-3_14. Methods Mol Biol. 2021. PMID: 33649956 Review.
-
Utilization of dietary mixed-linkage β-glucans by the Firmicute Blautia producta.J Biol Chem. 2023 Jun;299(6):104806. doi: 10.1016/j.jbc.2023.104806. Epub 2023 May 11. J Biol Chem. 2023. PMID: 37172725 Free PMC article.
-
Next-generation prebiotic promotes selective growth of bifidobacteria, suppressing Clostridioides difficile.Gut Microbes. 2021 Jan-Dec;13(1):1973835. doi: 10.1080/19490976.2021.1973835. Gut Microbes. 2021. PMID: 34553672 Free PMC article.
-
Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression.J Biol Chem. 2013 Aug 30;288(35):25194-25206. doi: 10.1074/jbc.M113.484733. Epub 2013 Jul 10. J Biol Chem. 2013. PMID: 23843461 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Argnani A, Leer RJ, van Luijk N, Pouwels PH. 1996. A convenient and reproducible method to genetically transform bacteria of the genus Bifidobacterium. Microbiology 142:109–114 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

