Translational homeostasis via the mRNA cap-binding protein, eIF4E
- PMID: 22578813
- PMCID: PMC4085128
- DOI: 10.1016/j.molcel.2012.04.004
Translational homeostasis via the mRNA cap-binding protein, eIF4E
Abstract
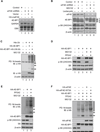
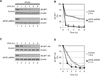
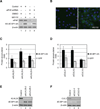
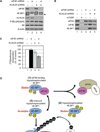
Translational control of gene expression plays a key role in many biological processes. Consequently, the activity of the translation apparatus is under tight homeostatic control. eIF4E, the mRNA 5' cap-binding protein, facilitates cap-dependent translation and is a major target for translational control. eIF4E activity is controlled by a family of repressor proteins, termed 4E-binding proteins (4E-BPs). Here, we describe the surprising finding that despite the importance of eIF4E for translation, a drastic knockdown of eIF4E caused only minor reduction in translation. This conundrum can be explained by the finding that 4E-BP1 is degraded in eIF4E-knockdown cells. Hypophosphorylated 4E-BP1, which binds to eIF4E, is degraded, whereas hyperphosphorylated 4E-BP1 is refractory to degradation. We identified the KLHL25-CUL3 complex as the E3 ubiquitin ligase, which targets hypophosphorylated 4E-BP1. Thus, the activity of eIF4E is under homeostatic control via the regulation of the levels of its repressor protein 4E-BP1 through ubiquitination.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures






Comment in
-
Translational homeostasis via eIF4E and 4E-BP1.Mol Cell. 2012 Jun 29;46(6):717-9. doi: 10.1016/j.molcel.2012.06.001. Mol Cell. 2012. PMID: 22749396
Similar articles
-
Characterizing the interaction of the mammalian eIF4E-related protein 4EHP with 4E-BP1.FEBS Lett. 2004 Apr 23;564(1-2):58-62. doi: 10.1016/S0014-5793(04)00313-8. FEBS Lett. 2004. PMID: 15094042
-
Translational repression by human 4E-BP1 in yeast specifically requires human eIF4E as target.J Biol Chem. 1999 Feb 5;274(6):3261-4. doi: 10.1074/jbc.274.6.3261. J Biol Chem. 1999. PMID: 9920863
-
RhoE inhibits 4E-BP1 phosphorylation and eIF4E function impairing cap-dependent translation.J Biol Chem. 2009 Dec 18;284(51):35287-96. doi: 10.1074/jbc.M109.050120. J Biol Chem. 2009. PMID: 19850923 Free PMC article.
-
Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): a master regulator of mRNA translation involved in tumorigenesis.Oncogene. 2016 Sep 8;35(36):4675-88. doi: 10.1038/onc.2015.515. Epub 2016 Feb 1. Oncogene. 2016. PMID: 26829052 Review.
-
4E-BP1, a multifactor regulated multifunctional protein.Cell Cycle. 2016;15(6):781-6. doi: 10.1080/15384101.2016.1151581. Cell Cycle. 2016. PMID: 26901143 Free PMC article. Review.
Cited by
-
PC-1/PrLZ confers resistance to rapamycin in prostate cancer cells through increased 4E-BP1 stability.Oncotarget. 2015 Aug 21;6(24):20356-69. doi: 10.18632/oncotarget.3931. Oncotarget. 2015. PMID: 26011939 Free PMC article.
-
What Is the Impact of mRNA 5' TL Heterogeneity on Translational Start Site Selection and the Mammalian Cellular Phenotype?Front Genet. 2016 Aug 31;7:156. doi: 10.3389/fgene.2016.00156. eCollection 2016. Front Genet. 2016. PMID: 27630668 Free PMC article. Review.
-
Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation.Mol Cell Biol. 2013 Jun;33(11):2285-301. doi: 10.1128/MCB.01517-12. Epub 2013 Apr 1. Mol Cell Biol. 2013. PMID: 23547259 Free PMC article.
-
Cap-dependent mRNA translation and the ubiquitin-proteasome system cooperate to promote ERBB2-dependent esophageal cancer phenotype.Cancer Gene Ther. 2012 Sep;19(9):609-18. doi: 10.1038/cgt.2012.39. Epub 2012 Jul 6. Cancer Gene Ther. 2012. PMID: 22767218 Free PMC article.
-
Active-site mTOR inhibitors augment HSV1-dICP0 infection in cancer cells via dysregulated eIF4E/4E-BP axis.PLoS Pathog. 2018 Aug 23;14(8):e1007264. doi: 10.1371/journal.ppat.1007264. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30138450 Free PMC article.
References
-
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. - PubMed
-
- Cormier P, Pyronnet S, Morales J, Mulner-Lorillon O, Sonenberg N, Belle R. eIF4E association with 4E-BP decreases rapidly following fertilization in sea urchin. Dev. Biol. 2001;232:275–283. - PubMed
-
- Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 1987;262:380–388. - PubMed
-
- Elia A, Constantinou C, Clemens MJ. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene. 2008;27:811–822. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

