Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins
- PMID: 22574288
- PMCID: PMC3345519
- DOI: 10.1016/j.celrep.2012.02.001
Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins
Abstract
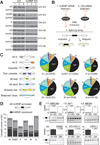
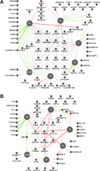
Understanding how RNA binding proteins control the splicing code is fundamental to human biology and disease. Here, we present a comprehensive study to elucidate how heterogeneous nuclear ribonucleoparticle (hnRNP) proteins, among the most abundant RNA binding proteins, coordinate to regulate alternative pre-mRNA splicing (AS) in human cells. Using splicing-sensitive microarrays, crosslinking and immunoprecipitation coupled with high-throughput sequencing (CLIP-seq), and cDNA sequencing, we find that more than half of all AS events are regulated by multiple hnRNP proteins and that some combinations of hnRNP proteins exhibit significant synergy, whereas others act antagonistically. Our analyses reveal position-dependent RNA splicing maps, in vivo consensus binding sites, a surprising level of cross- and autoregulation among hnRNP proteins, and the coordinated regulation by hnRNP proteins of dozens of other RNA binding proteins and genes associated with cancer. Our findings define an unprecedented degree of complexity and compensatory relationships among hnRNP proteins and their splicing targets that likely confer robustness to cells.
Figures




Similar articles
-
Transcriptome-wide RNA interaction profiling reveals physical and functional targets of hnRNP L in human T cells.Mol Cell Biol. 2014 Jan;34(1):71-83. doi: 10.1128/MCB.00740-13. Epub 2013 Oct 28. Mol Cell Biol. 2014. PMID: 24164894 Free PMC article.
-
Multiple and specific mRNA processing targets for the major human hnRNP proteins.Mol Cell Biol. 2008 Oct;28(19):6033-43. doi: 10.1128/MCB.00726-08. Epub 2008 Jul 21. Mol Cell Biol. 2008. PMID: 18644864 Free PMC article.
-
Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L.RNA Biol. 2014;11(2):146-55. doi: 10.4161/rna.27991. Epub 2014 Feb 7. RNA Biol. 2014. PMID: 24526010 Free PMC article.
-
hnRNP proteins and splicing control.Adv Exp Med Biol. 2007;623:123-47. doi: 10.1007/978-0-387-77374-2_8. Adv Exp Med Biol. 2007. PMID: 18380344 Review.
-
Splicing at the phase-separated nuclear speckle interface: a model.Nucleic Acids Res. 2021 Jan 25;49(2):636-645. doi: 10.1093/nar/gkaa1209. Nucleic Acids Res. 2021. PMID: 33337476 Free PMC article. Review.
Cited by
-
The long and the short of TRF2 in neurogenesis.Cell Cycle. 2016 Nov 16;15(22):3026-3032. doi: 10.1080/15384101.2016.1222339. Epub 2016 Aug 26. Cell Cycle. 2016. PMID: 27565210 Free PMC article. Review.
-
Cleavage and polyadenylation specificity factor 30: An RNA-binding zinc-finger protein with an unexpected 2Fe-2S cluster.Proc Natl Acad Sci U S A. 2016 Apr 26;113(17):4700-5. doi: 10.1073/pnas.1517620113. Epub 2016 Apr 11. Proc Natl Acad Sci U S A. 2016. PMID: 27071088 Free PMC article.
-
PTBP1 and PTBP2 Repress Nonconserved Cryptic Exons.Cell Rep. 2016 Sep 27;17(1):104-113. doi: 10.1016/j.celrep.2016.08.071. Cell Rep. 2016. PMID: 27681424 Free PMC article.
-
Differences in alternative splicing and their potential underlying factors between animals and plants.J Adv Res. 2024 Oct;64:83-98. doi: 10.1016/j.jare.2023.11.017. Epub 2023 Nov 20. J Adv Res. 2024. PMID: 37981087 Free PMC article. Review.
-
Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements.Cell. 2013 Jan 31;152(3):453-66. doi: 10.1016/j.cell.2012.12.023. Cell. 2013. PMID: 23374342 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

