Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair
- PMID: 22573372
- PMCID: PMC3391136
- DOI: 10.1074/jbc.M112.374447
Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair
Abstract
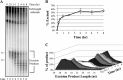
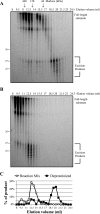
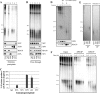
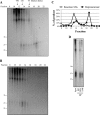
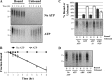
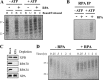
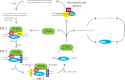
A wide range of environmental and carcinogenic agents form bulky lesions on DNA that are removed from the human genome in the form of short, ∼30-nucleotide oligonucleotides by the process of nucleotide excision repair. Although significant insights have been made regarding the mechanisms of damage recognition, dual incisions, and repair resynthesis during nucleotide excision repair, the fate of the dual incision/excision product is unknown. Using excision assays with both mammalian cell-free extract and purified proteins, we unexpectedly discovered that lesion-containing oligonucleotides are released from duplex DNA in complex with the general transcription and repair factor, Transcription Factor IIH (TFIIH). Release of excision products from TFIIH requires ATP but not ATP hydrolysis, and release occurs slowly, with a t(1/2) of 3.3 h. Excised oligonucleotides released from TFIIH then become bound by the single-stranded binding protein Replication Protein A or are targeted by cellular nucleases. These results provide a mechanism for release and an understanding of the initial fate of excised oligonucleotides during nucleotide excision repair.
Figures

Similar articles
-
Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo.J Biol Chem. 2013 Jul 19;288(29):20918-20926. doi: 10.1074/jbc.M113.482257. Epub 2013 Jun 8. J Biol Chem. 2013. PMID: 23749995 Free PMC article.
-
Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems.Methods Enzymol. 2006;408:189-213. doi: 10.1016/S0076-6879(06)08012-8. Methods Enzymol. 2006. PMID: 16793370
-
DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair.J Biol Chem. 2014 Sep 19;289(38):26574-26583. doi: 10.1074/jbc.M114.597088. Epub 2014 Aug 8. J Biol Chem. 2014. PMID: 25107903 Free PMC article.
-
Molecular basis for damage recognition and verification by XPC-RAD23B and TFIIH in nucleotide excision repair.DNA Repair (Amst). 2018 Nov;71:33-42. doi: 10.1016/j.dnarep.2018.08.005. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30174301 Free PMC article. Review.
-
Detection of the Excised, Damage-containing Oligonucleotide Products of Nucleotide Excision Repair in Human Cells.Photochem Photobiol. 2017 Jan;93(1):192-198. doi: 10.1111/php.12638. Epub 2016 Nov 3. Photochem Photobiol. 2017. PMID: 27634428 Free PMC article. Review.
Cited by
-
Nucleotide Excision Repair: Finely Tuned Molecular Orchestra of Early Pre-incision Events.Photochem Photobiol. 2017 Jan;93(1):166-177. doi: 10.1111/php.12647. Epub 2016 Nov 17. Photochem Photobiol. 2017. PMID: 27696486 Free PMC article. Review.
-
Spironolactone and XPB: An Old Drug with a New Molecular Target.Biomolecules. 2020 May 13;10(5):756. doi: 10.3390/biom10050756. Biomolecules. 2020. PMID: 32414008 Free PMC article. Review.
-
Life and death of circulating cell-free DNA.Cancer Biol Ther. 2019;20(8):1057-1067. doi: 10.1080/15384047.2019.1598759. Epub 2019 Apr 16. Cancer Biol Ther. 2019. PMID: 30990132 Free PMC article. Review.
-
Fluorescence detection of cellular nucleotide excision repair of damaged DNA.Sci Rep. 2014 Jul 4;4:5578. doi: 10.1038/srep05578. Sci Rep. 2014. PMID: 24993089 Free PMC article.
-
Remarkable Enhancement of Nucleotide Excision Repair of a Bulky Guanine Lesion in a Covalently Closed Circular DNA Plasmid Relative to the Same Linearized Plasmid.Biochemistry. 2020 Aug 11;59(31):2842-2848. doi: 10.1021/acs.biochem.0c00441. Epub 2020 Aug 2. Biochemistry. 2020. PMID: 32786887 Free PMC article.
References
-
- Huang J. C., Sancar A. (1994) Determination of minimum substrate size for human excinuclease. J. Biol. Chem. 269, 19034–19040 - PubMed
-
- Svoboda D. L., Taylor J. S., Hearst J. E., Sancar A. (1993) DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J. Biol. Chem. 268, 1931–1936 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

