The discrepancy between chromatin factor location and effect
- PMID: 22572961
- PMCID: PMC3414396
- DOI: 10.4161/nucl.19513
The discrepancy between chromatin factor location and effect
Abstract
The influence of chromatin on many cellular processes is well appreciated. Much has been learned by studying the role of chromatin remodeling and modifying complexes on individual genes. The seemingly straightforward models that inevitably arise from such studies are challenged by genome-wide analyses. Two recent studies in Saccharomyces cerevisiae provide unprecedented coverage of both the genome-wide location and the effect on gene expression for the majority of chromatin factors. Comparison of the overlap between location and expression effects reveals a large disconnect, with on average only 2.5% of occupied genes showing changes in expression. It is also interesting that only 24% of all expression effects are associated with chromatin factor occupancy. The large difference between location and effect likely reflects general properties inherent to regulation of gene expression through chromatin in yeast. Explanations for the discrepancy include gene-specific properties that exert a requirement for certain factors only on specific genes, as well as functional redundancy, whereby loss of a particular factor is compensated by others that function in a distinct but nevertheless compensatory manner. Since the majority of chromatin factor perturbations do show significant effects on specific subsets of genes, this implies the presence of different types of gene-specific properties that determine which chromatin factors a particular gene requires for proper expression. Understanding these gene-specific properties should be the focus of future studies aimed at understanding regulation of gene expression through chromatin.
Figures
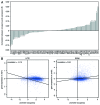
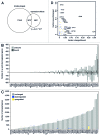
Comment on
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–49. doi: 10.1016/j.molcel.2011.03.026.
Similar articles
-
Mechanisms that specify promoter nucleosome location and identity.Cell. 2009 May 1;137(3):445-58. doi: 10.1016/j.cell.2009.02.043. Cell. 2009. PMID: 19410542 Free PMC article.
-
Tup1 stabilizes promoter nucleosome positioning and occupancy at transcriptionally plastic genes.Nucleic Acids Res. 2011 Nov 1;39(20):8803-19. doi: 10.1093/nar/gkr557. Epub 2011 Jul 23. Nucleic Acids Res. 2011. PMID: 21785133 Free PMC article.
-
[Chromatin structure and transcription regulation in Saccharomyces cerevisiae].Mol Biol (Mosk). 2010 Nov-Dec;44(6):966-79. Mol Biol (Mosk). 2010. PMID: 21290820 Russian.
-
The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae.Biochem Cell Biol. 2006 Aug;84(4):477-89. doi: 10.1139/o06-079. Biochem Cell Biol. 2006. PMID: 16936821 Review.
-
Genome-wide patterns of histone modifications in yeast.Nat Rev Mol Cell Biol. 2006 Sep;7(9):657-66. doi: 10.1038/nrm1986. Epub 2006 Aug 16. Nat Rev Mol Cell Biol. 2006. PMID: 16912715 Review.
Cited by
-
The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription.Genes Dev. 2014 Sep 15;28(18):1999-2012. doi: 10.1101/gad.250225.114. Genes Dev. 2014. PMID: 25228644 Free PMC article.
-
Coactivators and general transcription factors have two distinct dynamic populations dependent on transcription.EMBO J. 2017 Sep 15;36(18):2710-2725. doi: 10.15252/embj.201696035. Epub 2017 Jul 19. EMBO J. 2017. PMID: 28724529 Free PMC article.
-
Bridge-induced chromosome translocation in yeast relies upon a Rad54/Rdh54-dependent, Pol32-independent pathway.PLoS One. 2013 Apr 17;8(4):e60926. doi: 10.1371/journal.pone.0060926. Print 2013. PLoS One. 2013. PMID: 23613757 Free PMC article.
-
Global role for coactivator complexes in RNA polymerase II transcription.Transcription. 2019 Feb;10(1):29-36. doi: 10.1080/21541264.2018.1521214. Epub 2018 Oct 9. Transcription. 2019. PMID: 30299209 Free PMC article. Review.
-
The SAGA/TREX-2 subunit Sus1 binds widely to transcribed genes and affects mRNA turnover globally.Epigenetics Chromatin. 2018 Mar 29;11(1):13. doi: 10.1186/s13072-018-0184-2. Epigenetics Chromatin. 2018. PMID: 29598828 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
