Emerging Views on the CTD Code
- PMID: 22567385
- PMCID: PMC3335543
- DOI: 10.1155/2012/347214
Emerging Views on the CTD Code
Abstract
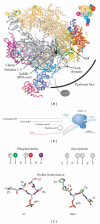
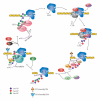
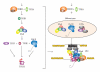
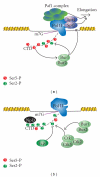
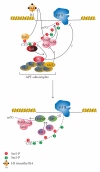
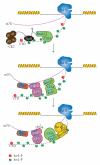
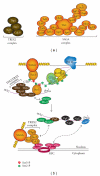
The C-terminal domain (CTD) of RNA polymerase II (Pol II) consists of conserved heptapeptide repeats that function as a binding platform for different protein complexes involved in transcription, RNA processing, export, and chromatin remodeling. The CTD repeats are subject to sequential waves of posttranslational modifications during specific stages of the transcription cycle. These patterned modifications have led to the postulation of the "CTD code" hypothesis, where stage-specific patterns define a spatiotemporal code that is recognized by the appropriate interacting partners. Here, we highlight the role of CTD modifications in directing transcription initiation, elongation, and termination. We examine the major readers, writers, and erasers of the CTD code and examine the relevance of describing patterns of posttranslational modifications as a "code." Finally, we discuss major questions regarding the function of the newly discovered CTD modifications and the fundamental insights into transcription regulation that will necessarily emerge upon addressing those challenges.
Figures
Similar articles
-
Removing quote marks from the RNA polymerase II CTD 'code'.Biosystems. 2021 Sep;207:104468. doi: 10.1016/j.biosystems.2021.104468. Epub 2021 Jun 30. Biosystems. 2021. PMID: 34216714
-
The CTD code of RNA polymerase II: a structural view.Wiley Interdiscip Rev RNA. 2013 Jan-Feb;4(1):1-16. doi: 10.1002/wrna.1138. Epub 2012 Oct 5. Wiley Interdiscip Rev RNA. 2013. PMID: 23042580 Review.
-
Simplicity is the Ultimate Sophistication-Crosstalk of Post-translational Modifications on the RNA Polymerase II.J Mol Biol. 2021 Jul 9;433(14):166912. doi: 10.1016/j.jmb.2021.166912. Epub 2021 Mar 5. J Mol Biol. 2021. PMID: 33676925 Free PMC article. Review.
-
Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner.Proc Natl Acad Sci U S A. 2017 May 16;114(20):E3944-E3953. doi: 10.1073/pnas.1700128114. Epub 2017 May 2. Proc Natl Acad Sci U S A. 2017. PMID: 28465432 Free PMC article.
-
The Ser7 of RNA Pol II-CTD influences the recruitment of Cdc73 for mRNA transcription.Int J Biol Macromol. 2024 Jan;254(Pt 2):127881. doi: 10.1016/j.ijbiomac.2023.127881. Epub 2023 Nov 8. Int J Biol Macromol. 2024. PMID: 37944716
Cited by
-
Engineered Covalent Inactivation of TFIIH-Kinase Reveals an Elongation Checkpoint and Results in Widespread mRNA Stabilization.Mol Cell. 2016 Aug 4;63(3):433-44. doi: 10.1016/j.molcel.2016.06.036. Epub 2016 Jul 28. Mol Cell. 2016. PMID: 27477907 Free PMC article.
-
Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae.Bioessays. 2015 Jun;37(6):666-77. doi: 10.1002/bies.201400220. Epub 2015 Mar 20. Bioessays. 2015. PMID: 25801414 Free PMC article. Review.
-
Cdc15 Phosphorylates the C-terminal Domain of RNA Polymerase II for Transcription during Mitosis.J Biol Chem. 2017 Mar 31;292(13):5507-5518. doi: 10.1074/jbc.M116.761056. Epub 2017 Feb 15. J Biol Chem. 2017. PMID: 28202544 Free PMC article.
-
Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation.J Biol Chem. 2012 Nov 9;287(46):38755-66. doi: 10.1074/jbc.M112.412015. Epub 2012 Oct 1. J Biol Chem. 2012. PMID: 23027873 Free PMC article.
-
Ash2L enables P53-dependent apoptosis by favoring stable transcription pre-initiation complex formation on its pro-apoptotic target promoters.Oncogene. 2015 May 7;34(19):2461-70. doi: 10.1038/onc.2014.198. Epub 2014 Jul 14. Oncogene. 2015. PMID: 25023704 Free PMC article.
References
-
- Cramer P, Armache KJ, Baumli S, et al. Structure of eukaryotic RNA polymerases. Annual Review of Biophysics. 2008;37:337–352. - PubMed
-
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes & Development. 2003;17(14):1691–1702. - PubMed
-
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends in Genetics. 2007;23(12):614–622. - PubMed
-
- Werner M, Thuriaux P, Soutourina J. Structure-function analysis of RNA polymerases I and III. Current Opinion in Structural Biology. 2009;19(6):740–745. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

