Transcriptional repression of ATF4 gene by CCAAT/enhancer-binding protein β (C/EBPβ) differentially regulates integrated stress response
- PMID: 22556424
- PMCID: PMC3381154
- DOI: 10.1074/jbc.M112.351783
Transcriptional repression of ATF4 gene by CCAAT/enhancer-binding protein β (C/EBPβ) differentially regulates integrated stress response
Abstract
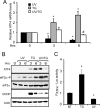
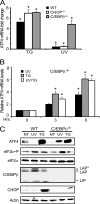
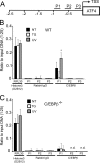
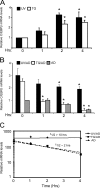
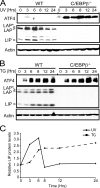
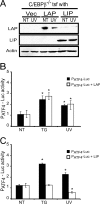
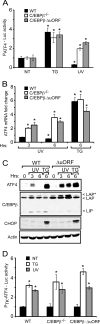
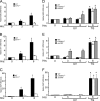
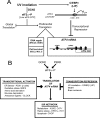
Different environmental stresses induce the phosphorylation of eIF2 (eIF2∼P), repressing global protein synthesis coincident with preferential translation of ATF4. ATF4 is a transcriptional activator of genes involved in metabolism and nutrient uptake, antioxidation, and regulation of apoptosis. Because ATF4 is a common downstream target that integrates signaling from different eIF2 kinases and their respective stress signals, the eIF2∼P/ATF4 pathway is collectively referred to as the integrated stress response. Although eIF2∼P elicits translational control in response to many different stresses, there are selected stresses, such as exposure to UV irradiation, that do not increase ATF4 expression despite robust eIF2∼P. The rationale for this discordant induction of ATF4 expression and eIF2∼P in response to UV irradiation is that transcription of ATF4 is repressed, and therefore ATF4 mRNA is not available for preferential translation. In this study, we show that C/EBPβ is a transcriptional repressor of ATF4 during UV stress. C/EBPβ binds to critical elements in the ATF4 promoter, resulting in its transcriptional repression. Expression of C/EBPβ increases in response to UV stress, and the liver-enriched inhibitory protein (LIP) isoform of C/EBPβ, but not the liver-enriched activating protein (LAP) version, represses ATF4 transcription. Loss of the liver-enriched inhibitory protein isoform results in increased ATF4 mRNA levels in response to UV irradiation and subsequent recovery of ATF4 translation, leading to enhanced expression of its target genes. Together these results illustrate how eIF2∼P and translational control combined with transcription factors regulated by alternative signaling pathways can direct programs of gene expression that are specifically tailored to each environmental stress.
Figures

Similar articles
-
Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response.J Biol Chem. 2010 Oct 22;285(43):33165-33174. doi: 10.1074/jbc.M110.167213. Epub 2010 Aug 23. J Biol Chem. 2010. PMID: 20732869 Free PMC article.
-
CCAAT/enhancer-binding protein β promotes receptor activator of nuclear factor-kappa-B ligand (RANKL) expression and osteoclast formation in the synovium in rheumatoid arthritis.Arthritis Res Ther. 2015 Feb 17;17(1):31. doi: 10.1186/s13075-015-0532-6. Arthritis Res Ther. 2015. PMID: 25811130 Free PMC article.
-
Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions.J Biol Chem. 2008 Mar 14;283(11):7064-73. doi: 10.1074/jbc.M708530200. Epub 2008 Jan 14. J Biol Chem. 2008. PMID: 18195013
-
Coping with stress: eIF2 kinases and translational control.Biochem Soc Trans. 2006 Feb;34(Pt 1):7-11. doi: 10.1042/BST20060007. Biochem Soc Trans. 2006. PMID: 16246168 Review.
-
Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism.Adv Nutr. 2012 May 1;3(3):307-21. doi: 10.3945/an.112.002113. Adv Nutr. 2012. PMID: 22585904 Free PMC article. Review.
Cited by
-
The integrated stress response.EMBO Rep. 2016 Oct;17(10):1374-1395. doi: 10.15252/embr.201642195. Epub 2016 Sep 14. EMBO Rep. 2016. PMID: 27629041 Free PMC article. Review.
-
Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR.Sci Adv. 2021 May 26;7(22):eabf0971. doi: 10.1126/sciadv.abf0971. Print 2021 May. Sci Adv. 2021. PMID: 34039602 Free PMC article.
-
TGR5 reduces macrophage migration through mTOR-induced C/EBPβ differential translation.J Clin Invest. 2014 Dec;124(12):5424-36. doi: 10.1172/JCI76289. Epub 2014 Nov 3. J Clin Invest. 2014. PMID: 25365223 Free PMC article.
-
Human Keratinocyte Differentiation Requires Translational Control by the eIF2α Kinase GCN2.J Invest Dermatol. 2017 Sep;137(9):1924-1934. doi: 10.1016/j.jid.2017.04.029. Epub 2017 May 17. J Invest Dermatol. 2017. PMID: 28528168 Free PMC article.
-
The Neuronal and Peripheral Expressed Membrane-Bound UNC93A Respond to Nutrient Availability in Mice.Front Mol Neurosci. 2017 Oct 31;10:351. doi: 10.3389/fnmol.2017.00351. eCollection 2017. Front Mol Neurosci. 2017. PMID: 29163028 Free PMC article.
References
-
- Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 - PubMed
-
- Wek R. C., Jiang H. Y., Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 - PubMed
-
- Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

