Protein kinase Cδ oxidation contributes to ERK inactivation in lupus T cells
- PMID: 22549474
- PMCID: PMC3414679
- DOI: 10.1002/art.34503
Protein kinase Cδ oxidation contributes to ERK inactivation in lupus T cells
Erratum in
- Arthritis Rheum. 2014 Mar;66(3):769. Patel, Dipak R [added]
Abstract
Objective: CD4+ T cells from patients with active lupus have impaired ERK pathway signaling that decreases DNA methyltransferase expression, resulting in DNA demethylation, overexpression of immune genes, and autoimmunity. The ERK pathway defect is due to impaired phosphorylation of T(505) in the protein kinase Cδ (PKCδ) activation loop. However, the mechanisms that prevent PKCδ T(505) phosphorylation in lupus T cells are unknown. Others have reported that oxidative modifications, and nitration in particular, of T cells as well as serum proteins correlate with lupus disease activity. We undertook this study to test our hypothesis that nitration inactivates PKCδ, contributing to impaired ERK pathway signaling in lupus T cells.
Methods: CD4+ T cells were purified from lupus patients and controls and then stimulated with phorbol myristate acetate (PMA). Signaling protein levels, nitration, and phosphorylation were quantitated by immunoprecipitation and immunoblotting of T cell lysates. Transfections were performed by electroporation.
Results: Treating CD4+ T cells with peroxynitrite nitrated PKCδ, preventing PKCδ T(505) phosphorylation and inhibiting ERK pathway signaling similar to that observed in lupus T cells. Patients with active lupus had higher nitrated T cell PKCδ levels than did controls, which correlated directly with disease activity, and antinitrotyrosine immunoprecipitations demonstrated that nitrated PKCδ, but not unmodified PKCδ, was refractory to PMA-stimulated T(505) phosphorylation, similar to PKCδ in peroxynitrite-treated cells.
Conclusion: Oxidative stress causes PKCδ nitration, which prevents its phosphorylation and contributes to the decreased ERK signaling in lupus T cells. These results identify PKCδ as a link between oxidative stress and the T cell epigenetic modifications in lupus.
Copyright © 2012 by the American College of Rheumatology.
Conflict of interest statement
Financial conflict of interest: There is no financial conflict of interest to disclose.
Figures
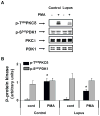

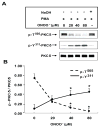
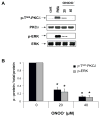
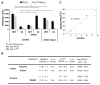
 ) in the supernatant (spnt) and the precipitate (pp) using CD4+ T cells from healthy donors and patients with active disease. * p≤0.01, ** p≤0.04, #≤0.02. Values are the mean ± SEM of six independent experiments. B: The table shows nitrated PKCδ (total PKCδ content in pp) and p-T505PKC δ (p-PKCδ in spnt + pp) as percent of total PKCδ (expressed in arbitrary units) in spnt and pp in each experimental condition. Values are the mean ± SEM of six experiments. * p values vs control. C: The graph shows the correlation between the nitrated PKCδ levels in lupus patients and the SLEDAI scores. p value was determined by analysis of variance (ANOVA).
) in the supernatant (spnt) and the precipitate (pp) using CD4+ T cells from healthy donors and patients with active disease. * p≤0.01, ** p≤0.04, #≤0.02. Values are the mean ± SEM of six independent experiments. B: The table shows nitrated PKCδ (total PKCδ content in pp) and p-T505PKC δ (p-PKCδ in spnt + pp) as percent of total PKCδ (expressed in arbitrary units) in spnt and pp in each experimental condition. Values are the mean ± SEM of six experiments. * p values vs control. C: The graph shows the correlation between the nitrated PKCδ levels in lupus patients and the SLEDAI scores. p value was determined by analysis of variance (ANOVA).Similar articles
-
Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus.J Immunol. 2007 Oct 15;179(8):5553-63. doi: 10.4049/jimmunol.179.8.5553. J Immunol. 2007. PMID: 17911642
-
T cell PKCδ kinase inactivation induces lupus-like autoimmunity in mice.Clin Immunol. 2015 Jun;158(2):193-203. doi: 10.1016/j.clim.2015.03.017. Epub 2015 Mar 28. Clin Immunol. 2015. PMID: 25829232 Free PMC article.
-
Oxidative stress, T cell DNA methylation, and lupus.Arthritis Rheumatol. 2014 Jun;66(6):1574-82. doi: 10.1002/art.38427. Arthritis Rheumatol. 2014. PMID: 24577881 Free PMC article.
-
Key role of ERK pathway signaling in lupus.Autoimmunity. 2010 Feb;43(1):17-22. doi: 10.3109/08916930903374832. Autoimmunity. 2010. PMID: 19961364 Free PMC article. Review.
-
Aberrant T cell ERK pathway signaling and chromatin structure in lupus.Autoimmun Rev. 2009 Jan;8(3):196-8. doi: 10.1016/j.autrev.2008.07.043. Epub 2008 Aug 22. Autoimmun Rev. 2009. PMID: 18723128 Free PMC article. Review.
Cited by
-
Environmental exposures, epigenetic changes and the risk of lupus.Lupus. 2014 May;23(6):568-76. doi: 10.1177/0961203313499419. Lupus. 2014. PMID: 24763540 Free PMC article. Review.
-
Environment and systemic autoimmune rheumatic diseases: an overview and future directions.Front Immunol. 2024 Sep 10;15:1456145. doi: 10.3389/fimmu.2024.1456145. eCollection 2024. Front Immunol. 2024. PMID: 39318630 Free PMC article. Review.
-
Oxidative Stress Contributes to Inflammatory and Cellular Damage in Systemic Lupus Erythematosus: Cellular Markers and Molecular Mechanism.J Inflamm Res. 2023 Feb 4;16:453-465. doi: 10.2147/JIR.S399284. eCollection 2023. J Inflamm Res. 2023. PMID: 36761905 Free PMC article. Review.
-
Epigenetically Altered T Cells Contribute to Lupus Flares.Cells. 2019 Feb 5;8(2):127. doi: 10.3390/cells8020127. Cells. 2019. PMID: 30764520 Free PMC article. Review.
-
The Role of Oxidative Stress in Epigenetic Changes Underlying Autoimmunity.Antioxid Redox Signal. 2022 Mar;36(7-9):423-440. doi: 10.1089/ars.2021.0066. Epub 2022 Jan 4. Antioxid Redox Signal. 2022. PMID: 34544258 Free PMC article. Review.
References
-
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–83. - PubMed
-
- Deng C, Yang J, Scott J, Hanash S, Richardson BC. Role of the ras-MAPK signaling pathway in the DNA methyltransferase response to DNA hypomethylation. Biol Chem. 1998;379(8–9):1113–20. - PubMed
-
- MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270(19):11327–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG025877/AG/NIA NIH HHS/United States
- R01 AR042525-18/AR/NIAMS NIH HHS/United States
- R01 AG025877-05/AG/NIA NIH HHS/United States
- P30 AR048310-10/AR/NIAMS NIH HHS/United States
- P30 ES017885-03/ES/NIEHS NIH HHS/United States
- P30 ES017885/ES/NIEHS NIH HHS/United States
- P30 AR048310/AR/NIAMS NIH HHS/United States
- AR-42525/AR/NIAMS NIH HHS/United States
- ES-015214/ES/NIEHS NIH HHS/United States
- R01 ES015214-04/ES/NIEHS NIH HHS/United States
- P30-ES-017885/ES/NIEHS NIH HHS/United States
- R01 ES015214/ES/NIEHS NIH HHS/United States
- P30-AR-048310/AR/NIAMS NIH HHS/United States
- R01 AR042525/AR/NIAMS NIH HHS/United States
- AG-25877/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

