The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output
- PMID: 22547686
- PMCID: PMC3434493
- DOI: 10.1128/MCB.00182-12
The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output
Abstract
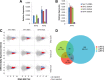
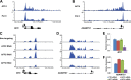
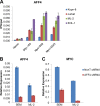
The elongation stage of transcription is highly regulated in metazoans. We previously purified the AFF1- and AFF4-containing super elongation complex (SEC) as a major regulator of development and cancer pathogenesis. Here, we report the biochemical isolation of SEC-like 2 (SEC-L2) and SEC-like 3 (SEC-L3) containing AFF2 and AFF3 in association with P-TEFb, ENL/MLLT1, and AF9/MLLT3. The SEC family members demonstrate high levels of polymerase II (Pol II) C-terminal domain kinase activity; however, only SEC is required for the proper induction of the HSP70 gene upon stress. Genome-wide mRNA-Seq analyses demonstrated that SEC-L2 and SEC-L3 control the expression of different subsets of genes, while AFF4/SEC plays a more dominant role in rapid transcriptional induction in cells. MYC is one of the direct targets of AFF4/SEC, and SEC recruitment to the MYC gene regulates its expression in different cancer cells, including those in acute myeloid or lymphoid leukemia. These findings suggest that AFF4/SEC could be a potential therapeutic target for the treatment of leukemia or other cancers associated with MYC overexpression.
Figures



Similar articles
-
Super elongation complex promotes early HIV transcription and its function is modulated by P-TEFb.Transcription. 2017 May 27;8(3):133-149. doi: 10.1080/21541264.2017.1295831. Epub 2017 Feb 17. Transcription. 2017. PMID: 28340332 Free PMC article.
-
Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):E636-45. doi: 10.1073/pnas.1107107108. Epub 2011 Aug 22. Proc Natl Acad Sci U S A. 2011. PMID: 21873227 Free PMC article.
-
AFF4 globally affects the release of paused RNA polymerase II in HEL cells.Yi Chuan. 2023 Aug 20;45(8):658-668. doi: 10.16288/j.yczz.23-143. Yi Chuan. 2023. PMID: 37609817
-
The super elongation complex (SEC) family in transcriptional control.Nat Rev Mol Cell Biol. 2012 Sep;13(9):543-7. doi: 10.1038/nrm3417. Epub 2012 Aug 16. Nat Rev Mol Cell Biol. 2012. PMID: 22895430 Review.
-
Collaboration of MLLT1/ENL, Polycomb and ATM for transcription and genome integrity.Nucleus. 2016 Apr 25;7(2):138-45. doi: 10.1080/19491034.2016.1177681. Nucleus. 2016. PMID: 27310306 Free PMC article. Review.
Cited by
-
Autophagy mediates proteolysis of NPM1 and HEXIM1 and sensitivity to BET inhibition in AML cells.Oncotarget. 2016 Nov 15;7(46):74917-74930. doi: 10.18632/oncotarget.12493. Oncotarget. 2016. PMID: 27732946 Free PMC article.
-
7SKiing on chromatin: Move globally, act locally.RNA Biol. 2016 Jun 2;13(6):545-53. doi: 10.1080/15476286.2016.1181254. Epub 2016 Apr 29. RNA Biol. 2016. PMID: 27128603 Free PMC article. Review.
-
Wnt/β-catenin signaling pathway in the tumor progression of adrenocortical carcinoma.Front Endocrinol (Lausanne). 2024 Jan 9;14:1260701. doi: 10.3389/fendo.2023.1260701. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38269250 Free PMC article. Review.
-
AF4 and AF4N protein complexes: recruitment of P-TEFb kinase, their interactome and potential functions.Am J Blood Res. 2015 Jun 15;5(1):10-24. eCollection 2015. Am J Blood Res. 2015. PMID: 26171280 Free PMC article.
-
Cat and Mouse: HIV Transcription in Latency, Immune Evasion and Cure/Remission Strategies.Viruses. 2019 Mar 18;11(3):269. doi: 10.3390/v11030269. Viruses. 2019. PMID: 30889861 Free PMC article. Review.
References
-
- Bitoun E, Davies KE. 2005. The robotic mouse: unravelling the function of AF4 in the cerebellum. Cerebellum 4:250–260 - PubMed
-
- Bitoun E, Oliver PL, Davies KE. 2007. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 16:92–106 - PubMed
-
- Bursen A, Moritz S, Gaussmann A, Dingermann T, Marschalek R. 2004. Interaction of AF4 wild-type and AF4.MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene 23:6237–6249 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
