Absolute quantification of somatic DNA alterations in human cancer
- PMID: 22544022
- PMCID: PMC4383288
- DOI: 10.1038/nbt.2203
Absolute quantification of somatic DNA alterations in human cancer
Abstract
We describe a computational method that infers tumor purity and malignant cell ploidy directly from analysis of somatic DNA alterations. The method, named ABSOLUTE, can detect subclonal heterogeneity and somatic homozygosity, and it can calculate statistical sensitivity for detection of specific aberrations. We used ABSOLUTE to analyze exome sequencing data from 214 ovarian carcinoma tumor-normal pairs. This analysis identified both pervasive subclonal somatic point-mutations and a small subset of predominantly clonal and homozygous mutations, which were overrepresented in the tumor suppressor genes TP53 and NF1 and in a candidate tumor suppressor gene CDK12. We also used ABSOLUTE to infer absolute allelic copy-number profiles from 3,155 diverse cancer specimens, revealing that genome-doubling events are common in human cancer, likely occur in cells that are already aneuploid, and influence pathways of tumor progression (for example, with recessive inactivation of NF1 being less common after genome doubling). ABSOLUTE will facilitate the design of clinical sequencing studies and studies of cancer genome evolution and intra-tumor heterogeneity.
Figures
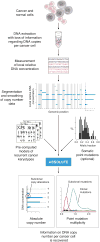

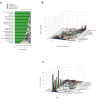
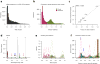

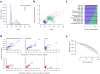
Comment in
-
Genetics: Understanding the ABSOLUTE genome.Nat Rev Clin Oncol. 2012 May 15;9(7):370. doi: 10.1038/nrclinonc.2012.86. Nat Rev Clin Oncol. 2012. PMID: 22585000 No abstract available.
-
ABSOLUTE cancer genomics.Nat Biotechnol. 2012 Jul 10;30(7):620-1. doi: 10.1038/nbt.2293. Nat Biotechnol. 2012. PMID: 22781683 Free PMC article.
Similar articles
-
Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations.Neoplasia. 2008 Dec;10(12):1362-72, following 1372. doi: 10.1593/neo.08784. Neoplasia. 2008. PMID: 19048115 Free PMC article.
-
A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal.PLoS Comput Biol. 2018 Feb 7;14(2):e1005965. doi: 10.1371/journal.pcbi.1005965. eCollection 2018 Feb. PLoS Comput Biol. 2018. PMID: 29415044 Free PMC article.
-
AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data.Bioinformatics. 2014 Apr 15;30(8):1056-1063. doi: 10.1093/bioinformatics/btt759. Epub 2014 Jan 2. Bioinformatics. 2014. PMID: 24389661 Free PMC article.
-
TP53 mutation-mediated genomic instability induces the evolution of chemoresistance and recurrence in epithelial ovarian cancer.Diagn Pathol. 2017 Feb 2;12(1):16. doi: 10.1186/s13000-017-0605-8. Diagn Pathol. 2017. PMID: 28148293 Free PMC article.
-
Somatic TP53 Mutations in the Era of Genome Sequencing.Cold Spring Harb Perspect Med. 2016 Nov 1;6(11):a026179. doi: 10.1101/cshperspect.a026179. Cold Spring Harb Perspect Med. 2016. PMID: 27503997 Free PMC article. Review.
Cited by
-
Genetic and microenvironmental intra-tumor heterogeneity impacts colorectal cancer evolution and metastatic development.Commun Biol. 2022 Sep 9;5(1):937. doi: 10.1038/s42003-022-03884-x. Commun Biol. 2022. PMID: 36085309 Free PMC article.
-
Knockout of immunotherapy prognostic marker genes eliminates the effect of the anti-PD-1 treatment.NPJ Precis Oncol. 2021 May 7;5(1):37. doi: 10.1038/s41698-021-00175-2. NPJ Precis Oncol. 2021. PMID: 33963274 Free PMC article.
-
GeTallele: A Method for Analysis of DNA and RNA Allele Frequency Distributions.Front Bioeng Biotechnol. 2020 Sep 16;8:1021. doi: 10.3389/fbioe.2020.01021. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33042959 Free PMC article.
-
Punctuated evolution of prostate cancer genomes.Cell. 2013 Apr 25;153(3):666-77. doi: 10.1016/j.cell.2013.03.021. Cell. 2013. PMID: 23622249 Free PMC article.
-
Long-term Benefit of PD-L1 Blockade in Lung Cancer Associated with JAK3 Activation.Cancer Immunol Res. 2015 Aug;3(8):855-63. doi: 10.1158/2326-6066.CIR-15-0024. Epub 2015 May 26. Cancer Immunol Res. 2015. PMID: 26014096 Free PMC article.
References
-
- Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. - PubMed
-
- Lindblad-Toh K, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat Biotechnol. 2000;18:1001–1005. - PubMed
-
- Zhao X, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08CA122833/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- 5R01 GM083299-14/GM/NIGMS NIH HHS/United States
- F32CA113126/CA/NCI NIH HHS/United States
- R01 GM083299/GM/NIGMS NIH HHS/United States
- U24CA143845/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U54 CA143798/CA/NCI NIH HHS/United States
- K08 CA122833/CA/NCI NIH HHS/United States
- U24CA143867/CA/NCI NIH HHS/United States
- F32 CA113126/CA/NCI NIH HHS/United States
- 5 T32 GM008313/GM/NIGMS NIH HHS/United States
- U24CA126546/CA/NCI NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
- U54CA143798/CA/NCI NIH HHS/United States
- U24 CA126546/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

