The 1000 Genomes Project: data management and community access
- PMID: 22543379
- PMCID: PMC3340611
- DOI: 10.1038/nmeth.1974
The 1000 Genomes Project: data management and community access
Abstract
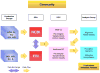
The 1000 Genomes Project was launched as one of the largest distributed data collection and analysis projects ever undertaken in biology. In addition to the primary scientific goals of creating both a deep catalog of human genetic variation and extensive methods to accurately discover and characterize variation using new sequencing technologies, the project makes all of its data publicly available. Members of the project data coordination center have developed and deployed several tools to enable widespread data access.
Figures
Similar articles
-
The International Genome Sample Resource (IGSR) collection of open human genomic variation resources.Nucleic Acids Res. 2020 Jan 8;48(D1):D941-D947. doi: 10.1093/nar/gkz836. Nucleic Acids Res. 2020. PMID: 31584097 Free PMC article.
-
The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data.Nucleic Acids Res. 2017 Jan 4;45(D1):D854-D859. doi: 10.1093/nar/gkw829. Epub 2016 Sep 15. Nucleic Acids Res. 2017. PMID: 27638885 Free PMC article.
-
Characterizing and interpreting genetic variation from personal genome sequencing.Methods Mol Biol. 2012;838:343-67. doi: 10.1007/978-1-61779-507-7_17. Methods Mol Biol. 2012. PMID: 22228021
-
Computational methods for discovering structural variation with next-generation sequencing.Nat Methods. 2009 Nov;6(11 Suppl):S13-20. doi: 10.1038/nmeth.1374. Nat Methods. 2009. PMID: 19844226 Review.
-
Pan-Genome Storage and Analysis Techniques.Methods Mol Biol. 2018;1704:29-53. doi: 10.1007/978-1-4939-7463-4_2. Methods Mol Biol. 2018. PMID: 29277862 Review.
Cited by
-
Unique genomic and neoepitope landscapes across tumors: a study across time, tissues, and space within a single lynch syndrome patient.Sci Rep. 2020 Jul 22;10(1):12190. doi: 10.1038/s41598-020-68939-7. Sci Rep. 2020. PMID: 32699259 Free PMC article.
-
Causal associations and potential mechanisms between inflammatory skin diseases and IgA nephropathy: a bi-directional Mendelian randomization study.Front Genet. 2024 Jul 25;15:1402302. doi: 10.3389/fgene.2024.1402302. eCollection 2024. Front Genet. 2024. PMID: 39119579 Free PMC article.
-
Origin, distribution, and function of three frequent coding polymorphisms in the gene for the human P2X7 ion channel.Front Pharmacol. 2022 Nov 18;13:1033135. doi: 10.3389/fphar.2022.1033135. eCollection 2022. Front Pharmacol. 2022. PMID: 36467077 Free PMC article.
-
Causal Relationships between Air Pollutant Exposure and Bone Mineral Density and the Risk of Bone Fractures: Evidence from a Two-Stage Mendelian Randomization Analysis.Toxics. 2023 Dec 30;12(1):27. doi: 10.3390/toxics12010027. Toxics. 2023. PMID: 38250984 Free PMC article.
-
Enhancing Discovery of Genetic Variants for Posttraumatic Stress Disorder Through Integration of Quantitative Phenotypes and Trauma Exposure Information.Biol Psychiatry. 2022 Apr 1;91(7):626-636. doi: 10.1016/j.biopsych.2021.09.020. Epub 2021 Sep 28. Biol Psychiatry. 2022. PMID: 34865855 Free PMC article.
References
-
- Baker M. Next-generation sequencing: adjusting to data overload. Nature Methods. 2010;7:495–499.
Publication types
MeSH terms
Grants and funding
- Z99 MH999999/ImNIH/Intramural NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- P20 MD006899/MD/NIMHD NIH HHS/United States
- P01 CA101937/CA/NCI NIH HHS/United States
- Z99 CL999999/ImNIH/Intramural NIH HHS/United States
- Z99 HL999999/ImNIH/Intramural NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 085532/WT_/Wellcome Trust/United Kingdom
- Z99 LM999999/ImNIH/Intramural NIH HHS/United States
- U01 HG006513/HG/NHGRI NIH HHS/United States
- G1000758/MRC_/Medical Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 095908/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

