Expression of mitochondrial non-coding RNAs (ncRNAs) is modulated by high risk human papillomavirus (HPV) oncogenes
- PMID: 22539350
- PMCID: PMC3375551
- DOI: 10.1074/jbc.M111.326694
Expression of mitochondrial non-coding RNAs (ncRNAs) is modulated by high risk human papillomavirus (HPV) oncogenes
Abstract
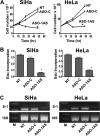
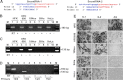
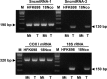
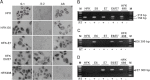
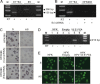
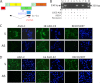
The study of RNA and DNA oncogenic viruses has proved invaluable in the discovery of key cellular pathways that are rendered dysfunctional during cancer progression. An example is high risk human papillomavirus (HPV), the etiological agent of cervical cancer. The role of HPV oncogenes in cellular immortalization and transformation has been extensively investigated. We reported the differential expression of a family of human mitochondrial non-coding RNAs (ncRNAs) between normal and cancer cells. Normal cells express a sense mitochondrial ncRNA (SncmtRNA) that seems to be required for cell proliferation and two antisense transcripts (ASncmtRNAs). In contrast, the ASncmtRNAs are down-regulated in cancer cells. To shed some light on the mechanisms that trigger down-regulation of the ASncmtRNAs, we studied human keratinocytes (HFK) immortalized with HPV. Here we show that immortalization of HFK with HPV-16 or 18 causes down-regulation of the ASncmtRNAs and induces the expression of a new sense transcript named SncmtRNA-2. Transduction of HFK with both E6 and E7 is sufficient to induce expression of SncmtRNA-2. Moreover, E2 oncogene is involved in down-regulation of the ASncmtRNAs. Knockdown of E2 in immortalized cells reestablishes in a reversible manner the expression of the ASncmtRNAs, suggesting that endogenous cellular factors(s) could play functions analogous to E2 during non-HPV-induced oncogenesis.
Figures

Similar articles
-
HPV-18 E2 protein downregulates antisense noncoding mitochondrial RNA-2, delaying replicative senescence of human keratinocytes.Aging (Albany NY). 2018 Dec 30;11(1):33-47. doi: 10.18632/aging.101711. Aging (Albany NY). 2018. PMID: 30595560 Free PMC article.
-
Down-regulation of the antisense mitochondrial non-coding RNAs (ncRNAs) is a unique vulnerability of cancer cells and a potential target for cancer therapy.J Biol Chem. 2014 Sep 26;289(39):27182-27198. doi: 10.1074/jbc.M114.558841. Epub 2014 Aug 6. J Biol Chem. 2014. PMID: 25100722 Free PMC article.
-
Human Papillomavirus E6/E7 and Long Noncoding RNA TMPOP2 Mutually Upregulated Gene Expression in Cervical Cancer Cells.J Virol. 2019 Apr 3;93(8):e01808-18. doi: 10.1128/JVI.01808-18. Print 2019 Apr 15. J Virol. 2019. PMID: 30728257 Free PMC article.
-
Effects of human papillomavirus (HPV) type 16 oncoproteins on the expression of involucrin in human keratinocytes.Virol J. 2012 Feb 14;9:36. doi: 10.1186/1743-422X-9-36. Virol J. 2012. PMID: 22333115 Free PMC article.
-
Post-Transcriptional Gene Regulation by HPV 16E6 and Its Host Protein Partners.Viruses. 2022 Jul 6;14(7):1483. doi: 10.3390/v14071483. Viruses. 2022. PMID: 35891463 Free PMC article. Review.
Cited by
-
Two RNAs or DNAs May Artificially Fuse Together at a Short Homologous Sequence (SHS) during Reverse Transcription or Polymerase Chain Reactions, and Thus Reporting an SHS-Containing Chimeric RNA Requires Extra Caution.PLoS One. 2016 May 5;11(5):e0154855. doi: 10.1371/journal.pone.0154855. eCollection 2016. PLoS One. 2016. PMID: 27148738 Free PMC article.
-
Functional implications of mitochondrial reactive oxygen species generated by oncogenic viruses.Front Biol (Beijing). 2014 Dec;9(6):423-436. doi: 10.1007/s11515-014-1332-0. Front Biol (Beijing). 2014. PMID: 25580106 Free PMC article.
-
It Is Imperative to Establish a Pellucid Definition of Chimeric RNA and to Clear Up a Lot of Confusion in the Relevant Research.Int J Mol Sci. 2017 Mar 28;18(4):714. doi: 10.3390/ijms18040714. Int J Mol Sci. 2017. PMID: 28350330 Free PMC article. Review.
-
RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication.PLoS Pathog. 2013;9(9):e1003610. doi: 10.1371/journal.ppat.1003610. Epub 2013 Sep 5. PLoS Pathog. 2013. PMID: 24039580 Free PMC article.
-
Different cancers, same target?Aging (Albany NY). 2017 Aug 8;9(8):1853-1854. doi: 10.18632/aging.101278. Aging (Albany NY). 2017. PMID: 28800297 Free PMC article. No abstract available.
References
-
- Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer. The next generation. Cell 144, 646–674 - PubMed
-
- D'Agostino D. M., Bernardi P., Chieco-Bianchi L., Ciminale V. (2005) Mitochondria as functional targets of proteins coded by human tumor viruses. Adv. Cancer Res. 94, 87–142 - PubMed
-
- zur Hausen H. (2009) Papillomaviruses in the causation of human cancers. A brief historical account. Virology 384, 260–265 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

