Interferon-inducible effector mechanisms in cell-autonomous immunity
- PMID: 22531325
- PMCID: PMC4150610
- DOI: 10.1038/nri3210
Interferon-inducible effector mechanisms in cell-autonomous immunity
Abstract
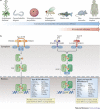
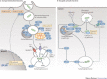
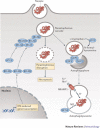
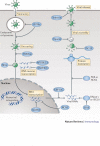
Interferons (IFNs) induce the expression of hundreds of genes as part of an elaborate antimicrobial programme designed to combat infection in all nucleated cells - a process termed cell-autonomous immunity. As described in this Review, recent genomic and subgenomic analyses have begun to assign functional properties to novel IFN-inducible effector proteins that restrict bacteria, protozoa and viruses in different subcellular compartments and at different stages of the pathogen life cycle. Several newly described host defence factors also participate in canonical oxidative and autophagic pathways by spatially coordinating their activities to enhance microbial killing. Together, these IFN-induced effector networks help to confer vertebrate host resistance to a vast and complex microbial world.
Conflict of interest statement
The author declares no competing financial interests.
Figures
Similar articles
-
Interferon-induced GTPases orchestrate host cell-autonomous defence against bacterial pathogens.Biochem Soc Trans. 2021 Jun 30;49(3):1287-1297. doi: 10.1042/BST20200900. Biochem Soc Trans. 2021. PMID: 34003245 Free PMC article. Review.
-
IFN-inducible GTPases and immunity to intracellular pathogens.Trends Immunol. 2004 Nov;25(11):601-9. doi: 10.1016/j.it.2004.08.010. Trends Immunol. 2004. PMID: 15489189 Review.
-
Importance of Type I and III Interferons at Respiratory and Intestinal Barrier Surfaces.Front Immunol. 2020 Dec 11;11:608645. doi: 10.3389/fimmu.2020.608645. eCollection 2020. Front Immunol. 2020. PMID: 33362795 Free PMC article. Review.
-
IFN-inducible GTPases in host cell defense.Cell Host Microbe. 2012 Oct 18;12(4):432-44. doi: 10.1016/j.chom.2012.09.007. Cell Host Microbe. 2012. PMID: 23084913 Free PMC article. Review.
-
Interferon-inducible GTPases in cell autonomous and innate immunity.Cell Microbiol. 2016 Feb;18(2):168-80. doi: 10.1111/cmi.12546. Epub 2015 Dec 28. Cell Microbiol. 2016. PMID: 26572694 Review.
Cited by
-
Cell-autonomous stress responses in innate immunity.J Leukoc Biol. 2017 Jan;101(1):77-86. doi: 10.1189/jlb.2MR0416-201R. Epub 2016 Oct 12. J Leukoc Biol. 2017. PMID: 27733577 Free PMC article. Review.
-
Type I interferons in infectious disease.Nat Rev Immunol. 2015 Feb;15(2):87-103. doi: 10.1038/nri3787. Nat Rev Immunol. 2015. PMID: 25614319 Free PMC article. Review.
-
In Vitro Modelling of Chlamydia trachomatis Infection in the Etiopathogenesis of Male Infertility and Reactive Arthritis.Front Cell Infect Microbiol. 2022 Jan 31;12:840802. doi: 10.3389/fcimb.2022.840802. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35174109 Free PMC article. Review.
-
Review of Immunologic Manifestations of COVID-19 Infection and Vaccination.Heart Fail Clin. 2023 Apr;19(2):177-184. doi: 10.1016/j.hfc.2022.08.006. Heart Fail Clin. 2023. PMID: 36863809 Free PMC article. Review.
-
Interferon and interferon-stimulated genes in HBV treatment.Front Immunol. 2022 Dec 1;13:1034968. doi: 10.3389/fimmu.2022.1034968. eCollection 2022. Front Immunol. 2022. PMID: 36531993 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

