Virion assembly factories in the nucleus of polyomavirus-infected cells
- PMID: 22496654
- PMCID: PMC3320610
- DOI: 10.1371/journal.ppat.1002630
Virion assembly factories in the nucleus of polyomavirus-infected cells
Abstract
Most DNA viruses replicate in the cell nucleus, although the specific sites of virion assembly are as yet poorly defined. Electron microscopy on freeze-substituted, plastic-embedded sections of murine polyomavirus (PyV)-infected 3T3 mouse fibroblasts or mouse embryonic fibroblasts (MEFs) revealed tubular structures in the nucleus adjacent to clusters of assembled virions, with virions apparently "shed" or "budding" from their ends. Promyelocytic leukemia nuclear bodies (PML-NBs) have been suggested as possible sites for viral replication of polyomaviruses (BKV and SV40), herpes simplex virus (HSV), and adenovirus (Ad). Immunohistochemistry and FISH demonstrated co-localization of the viral T-antigen (Tag), PyV DNA, and the host DNA repair protein MRE11, adjacent to the PML-NBs. In PML⁻/⁻ MEFs the co-localization of MRE11, Tag, and PyV DNA remained unchanged, suggesting that the PML protein itself was not responsible for their association. Furthermore, PyV-infected PML⁻/⁻ MEFs and PML⁻/⁻ mice replicated wild-type levels of infectious virus. Therefore, although the PML protein may identify sites of PyV replication, neither the observed "virus factories" nor virus assembly were dependent on PML. The ultrastructure of the tubes suggests a new model for the encapsidation of small DNA viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

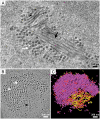

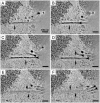
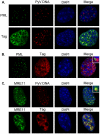
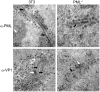
Similar articles
-
Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection.mBio. 2011 Feb 8;2(1):e00281-10. doi: 10.1128/mBio.00281-10. Print 2011. mBio. 2011. PMID: 21304169 Free PMC article.
-
Functional connection between Rad51 and PML in homology-directed repair.PLoS One. 2011;6(10):e25814. doi: 10.1371/journal.pone.0025814. Epub 2011 Oct 5. PLoS One. 2011. PMID: 21998700 Free PMC article.
-
Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells.J Virol. 2007 Sep;81(18):10123-36. doi: 10.1128/JVI.01009-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626080 Free PMC article.
-
Targeting promyelocytic leukemia protein: a means to regulating PML nuclear bodies.Int J Biol Sci. 2009 May 22;5(4):366-76. doi: 10.7150/ijbs.5.366. Int J Biol Sci. 2009. PMID: 19471587 Free PMC article. Review.
-
PML nuclear bodies as sites of epigenetic regulation.Front Biosci (Landmark Ed). 2009 Jan 1;14(4):1325-36. doi: 10.2741/3311. Front Biosci (Landmark Ed). 2009. PMID: 19273133 Review.
Cited by
-
Activation of DNA damage repair pathways by murine polyomavirus.Virology. 2016 Oct;497:346-356. doi: 10.1016/j.virol.2016.07.028. Epub 2016 Aug 16. Virology. 2016. PMID: 27529739 Free PMC article.
-
Replication Compartments of Eukaryotic and Bacterial DNA Viruses: Common Themes Between Different Domains of Host Cells.Annu Rev Virol. 2022 Sep 29;9(1):307-327. doi: 10.1146/annurev-virology-012822-125828. Annu Rev Virol. 2022. PMID: 36173697 Free PMC article. Review.
-
The Major Capsid Protein, VP1, of the Mouse Polyomavirus Stimulates the Activity of Tubulin Acetyltransferase 1 by Microtubule Stabilization.Viruses. 2020 Feb 18;12(2):227. doi: 10.3390/v12020227. Viruses. 2020. PMID: 32085463 Free PMC article.
-
JC virus inclusions in progressive multifocal leukoencephalopathy: scaffolding promyelocytic leukemia nuclear bodies grow with cell cycle transition through an S-to-G2-like state in enlarging oligodendrocyte nuclei.J Neuropathol Exp Neurol. 2014 May;73(5):442-53. doi: 10.1097/NEN.0000000000000066. J Neuropathol Exp Neurol. 2014. PMID: 24709678 Free PMC article.
-
Polyomavirus interaction with the DNA damage response.Virol Sin. 2015 Apr;30(2):122-9. doi: 10.1007/s12250-015-3583-6. Epub 2015 Apr 20. Virol Sin. 2015. PMID: 25910481 Free PMC article. Review.
References
-
- Wileman T. Aggresomes and Pericentriolar Sites of Virus Assembly: Cellular Defense or Viral Design? Annu Rev Microbiol. 2007;61:149–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

