Roles of Kruppel-associated Box (KRAB)-associated Co-repressor KAP1 Ser-473 Phosphorylation in DNA Damage Response
- PMID: 22496453
- PMCID: PMC3365928
- DOI: 10.1074/jbc.M111.313262
Roles of Kruppel-associated Box (KRAB)-associated Co-repressor KAP1 Ser-473 Phosphorylation in DNA Damage Response
Abstract
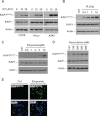
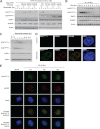
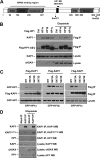
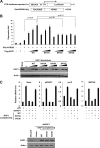
The Kruppel-associated box (KRAB)-associated co-repressor KAP1 is an essential nuclear co-repressor for the KRAB zinc finger protein superfamily of transcriptional factors. Ataxia telangiectasia mutated (ATM)-Chk2 and ATM- and Rad3-related (ATR)-Chk1 are two primary kinase signaling cascades activated in response to DNA damage. A growing body of evidence suggests that ATM and ATR phosphorylate KAP1 at Ser-824 in response to DNA damage and regulate KAP1-dependent chromatin condensation, DNA repair, and gene expression. Here, we show that, depending on the type of DNA damage that occurs, KAP1 Ser-473 can be phosphorylated by ATM-Chk2 or ATR-Chk1 kinases. Phosphorylation of KAP1 at Ser-473 attenuated its binding to the heterochromatin protein 1 family proteins and inhibited its transcriptional repression of KRAB-zinc finger protein (KRAB-ZFP) target genes. Moreover, KAP1 Ser-473 phosphorylation induced by DNA damage stimulated KAP1-E2F1 binding. Overexpression of heterochromatin protein 1 significantly inhibited E2F1-KAP1 binding. Elimination of KAP1 Ser-473 phosphorylation increased E2F1-targeted proapoptotic gene expression and E2F1-induced apoptosis in response to DNA damage. Furthermore, loss of phosphorylation of KAP1 Ser-473 led to less BRCA1 focus formation and slower kinetics of loss of γH2AX foci after DNA damage. KAP1 Ser-473 phosphorylation was required for efficient DNA repair and cell survival in response to DNA damage. Our studies reveal novel functions of KAP1 Ser-473 phosphorylation under stress.
Figures


Similar articles
-
Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression.J Biol Chem. 2007 Dec 14;282(50):36177-89. doi: 10.1074/jbc.M706912200. Epub 2007 Oct 17. J Biol Chem. 2007. PMID: 17942393
-
The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation.Mol Cancer Res. 2012 Mar;10(3):401-14. doi: 10.1158/1541-7786.MCR-11-0134. Epub 2011 Dec 28. Mol Cancer Res. 2012. PMID: 22205726 Free PMC article.
-
KAP1 depletion increases PML nuclear body number in concert with ultrastructural changes in chromatin.Cell Cycle. 2011 Jan 15;10(2):308-22. doi: 10.4161/cc.10.2.14551. Epub 2011 Jan 15. Cell Cycle. 2011. PMID: 21228624
-
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer.Adv Cancer Res. 2010;108:73-112. doi: 10.1016/B978-0-12-380888-2.00003-0. Adv Cancer Res. 2010. PMID: 21034966 Review.
-
SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins.Bioessays. 2005 Apr;27(4):397-407. doi: 10.1002/bies.20204. Bioessays. 2005. PMID: 15770685 Review.
Cited by
-
KRAB domain of ZFP568 disrupts TRIM28-mediated abnormal interactions in cancer cells.NAR Cancer. 2020 Jun;2(2):zcaa007. doi: 10.1093/narcan/zcaa007. Epub 2020 May 18. NAR Cancer. 2020. PMID: 32743551 Free PMC article.
-
Epigenetic Restriction Factors (eRFs) in Virus Infection.Viruses. 2024 Jan 25;16(2):183. doi: 10.3390/v16020183. Viruses. 2024. PMID: 38399958 Free PMC article. Review.
-
The role of tripartite motif-containing 28 in cancer progression and its therapeutic potentials.Front Oncol. 2023 Jan 23;13:1100134. doi: 10.3389/fonc.2023.1100134. eCollection 2023. Front Oncol. 2023. PMID: 36756159 Free PMC article. Review.
-
Human phospho-signaling networks of SARS-CoV-2 infection are rewired by population genetic variants.Mol Syst Biol. 2022 May;18(5):e10823. doi: 10.15252/msb.202110823. Mol Syst Biol. 2022. PMID: 35579274 Free PMC article.
-
KAP1 facilitates reinstatement of heterochromatin after DNA replication.Nucleic Acids Res. 2018 Sep 28;46(17):8788-8802. doi: 10.1093/nar/gky580. Nucleic Acids Res. 2018. PMID: 29955894 Free PMC article.
References
-
- Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067–2078 - PubMed
-
- Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R. (2000) Mice lacking the transcriptional corepressor TIF1β are defective in early postimplantation development. Development 127, 2955–2963 - PubMed
-
- Weber P., Cammas F., Gerard C., Metzger D., Chambon P., Losson R., Mark M. (2002) Germ cell expression of the transcriptional co-repressor TIF1β is required for the maintenance of spermatogenesis in the mouse. Development 129, 2329–2337 - PubMed
-
- Peng H., Begg G. E., Schultz D. C., Friedman J. R., Jensen D. E., Speicher D. W., Rauscher F. J., 3rd (2000) Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 295, 1139–1162 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

