Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly
- PMID: 22473132
- PMCID: PMC3364751
- DOI: 10.1038/emboj.2012.82
Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly
Abstract
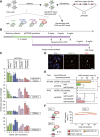
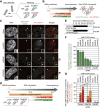
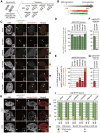
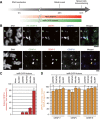
The kinetochore is responsible for accurate chromosome segregation. However, the mechanism by which kinetochores assemble and are maintained remains unclear. Here we report that de novo CENP-A assembly and kinetochore formation on human centromeric alphoid DNA arrays is regulated by a histone H3K9 acetyl/methyl balance. Tethering of histone acetyltransferases (HATs) to alphoid DNA arrays breaks a cell type-specific barrier for de novo stable CENP-A assembly and induces assembly of other kinetochore proteins at the ectopic alphoid site. Similar results are obtained following tethering of CENP-A deposition factors hMis18α or HJURP. HAT tethering bypasses the need for hMis18α, but HJURP is still required for de novo kinetochore assembly. In contrast, H3K9 methylation following tethering of H3K9 tri-methylase (Suv39h1) to the array prevents de novo CENP-A assembly and kinetochore formation. CENP-A arrays assembled de novo by this mechanism can form human artificial chromosomes (HACs) that are propagated indefinitely in human cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly.J Cell Sci. 2015 Dec 15;128(24):4572-87. doi: 10.1242/jcs.180786. Epub 2015 Nov 2. J Cell Sci. 2015. PMID: 26527398 Free PMC article.
-
KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation.Dev Cell. 2016 Jun 6;37(5):413-27. doi: 10.1016/j.devcel.2016.05.006. Dev Cell. 2016. PMID: 27270040 Free PMC article.
-
Genetic and epigenetic regulation of centromeres: a look at HAC formation.Chromosome Res. 2015 Feb;23(1):87-103. doi: 10.1007/s10577-015-9470-z. Chromosome Res. 2015. PMID: 25682171 Review.
-
HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore.J Cell Biol. 2011 Jul 25;194(2):229-43. doi: 10.1083/jcb.201012017. Epub 2011 Jul 18. J Cell Biol. 2011. PMID: 21768289 Free PMC article.
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
Cited by
-
Comparative study of artificial chromosome centromeres in human and murine cells.Eur J Hum Genet. 2013 Sep;21(9):948-56. doi: 10.1038/ejhg.2012.296. Epub 2013 Feb 13. Eur J Hum Genet. 2013. PMID: 23403904 Free PMC article.
-
Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution.Genome Biol. 2013 Jan 30;14(1):R10. doi: 10.1186/gb-2013-14-1-r10. Genome Biol. 2013. PMID: 23363705 Free PMC article.
-
A new assay for measuring chromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells.BMC Cancer. 2013 May 22;13:252. doi: 10.1186/1471-2407-13-252. BMC Cancer. 2013. PMID: 23694679 Free PMC article.
-
A portable BRCA1-HAC (human artificial chromosome) module for analysis of BRCA1 tumor suppressor function.Nucleic Acids Res. 2014 Dec 1;42(21):e164. doi: 10.1093/nar/gku870. Epub 2014 Sep 26. Nucleic Acids Res. 2014. PMID: 25260588 Free PMC article.
-
The centromere: chromatin foundation for the kinetochore machinery.Dev Cell. 2014 Sep 8;30(5):496-508. doi: 10.1016/j.devcel.2014.08.016. Dev Cell. 2014. PMID: 25203206 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

