Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5'-overhangs
- PMID: 22467875
- PMCID: PMC3370212
- DOI: 10.1074/jbc.M112.347278
Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5'-overhangs
Abstract
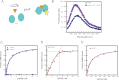
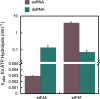
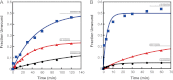
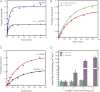
During eukaryotic translation initiation, the 43 S ribosomal pre-initiation complex is recruited to the 5'-end of an mRNA through its interaction with the 7-methylguanosine cap, and it subsequently scans along the mRNA to locate the start codon. Both mRNA recruitment and scanning require the removal of secondary structure within the mRNA. Eukaryotic translation initiation factor 4A is an essential component of the translational machinery thought to participate in the clearing of secondary structural elements in the 5'-untranslated regions of mRNAs. eIF4A is part of the 5'-7-methylguanosine cap-binding complex, eIF4F, along with eIF4E, the cap-binding protein, and the scaffolding protein eIF4G. Here, we show that Saccharomyces cerevisiae eIF4F has a strong preference for unwinding an RNA duplex with a single-stranded 5'-overhang versus the same duplex with a 3'-overhang or without an overhang. In contrast, eIF4A on its own has little RNA substrate specificity. Using a series of deletion constructs of eIF4G, we demonstrate that its three previously elucidated RNA binding domains work together to provide eIF4F with its 5'-end specificity, both by promoting unwinding of substrates with 5'-overhangs and inhibiting unwinding of substrates with 3'-overhangs. Our data suggest that the RNA binding domains of eIF4G provide the S. cerevisiae eIF4F complex with a second mechanism, in addition to the eIF4E-cap interaction, for directing the binding of pre-initiation complexes to the 5'-ends of mRNAs and for biasing scanning in the 5' to 3' direction.
Figures


Similar articles
-
Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity.Elife. 2017 Nov 30;6:e31476. doi: 10.7554/eLife.31476. Elife. 2017. PMID: 29192585 Free PMC article.
-
The eukaryotic initiation factor (eIF) 4G HEAT domain promotes translation re-initiation in yeast both dependent on and independent of eIF4A mRNA helicase.J Biol Chem. 2010 Jul 16;285(29):21922-33. doi: 10.1074/jbc.M110.132027. Epub 2010 May 12. J Biol Chem. 2010. PMID: 20463023 Free PMC article.
-
The domains of yeast eIF4G, eIF4E and the cap fine-tune eIF4A activities through an intricate network of stimulatory and inhibitory effects.Nucleic Acids Res. 2022 Jun 24;50(11):6497-6510. doi: 10.1093/nar/gkac437. Nucleic Acids Res. 2022. PMID: 35689631 Free PMC article.
-
Manipulation of the host translation initiation complex eIF4F by DNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1511-6. doi: 10.1042/BST0381511. Biochem Soc Trans. 2010. PMID: 21118117 Review.
-
eIF4F: a retrospective.J Biol Chem. 2015 Oct 2;290(40):24091-9. doi: 10.1074/jbc.R115.675280. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324716 Free PMC article. Review.
Cited by
-
An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation.Nucleic Acids Res. 2016 May 19;44(9):4368-80. doi: 10.1093/nar/gkw172. Epub 2016 Mar 16. Nucleic Acids Res. 2016. PMID: 26984530 Free PMC article.
-
Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains.RNA. 2013 Feb;19(2):191-207. doi: 10.1261/rna.035881.112. Epub 2012 Dec 12. RNA. 2013. PMID: 23236192 Free PMC article.
-
Toward a molecular understanding of RNA remodeling by DEAD-box proteins.RNA Biol. 2013 Jan;10(1):44-55. doi: 10.4161/rna.22210. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995827 Free PMC article. Review.
-
Translation initiation of alphavirus mRNA reveals new insights into the topology of the 48S initiation complex.Nucleic Acids Res. 2018 May 4;46(8):4176-4187. doi: 10.1093/nar/gky071. Nucleic Acids Res. 2018. PMID: 29415133 Free PMC article.
-
Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity.Elife. 2017 Nov 30;6:e31476. doi: 10.7554/eLife.31476. Elife. 2017. PMID: 29192585 Free PMC article.
References
-
- Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W. C., Sonenberg N. (2011) mRNA helicases. The tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12, 235–245 - PubMed
-
- Gorbalenya A. E., Koonin E. V. (1993) Helicases. Amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3, 419–429
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

