Design of Bcl-2 and Bcl-xL inhibitors with subnanomolar binding affinities based upon a new scaffold
- PMID: 22448988
- PMCID: PMC3397176
- DOI: 10.1021/jm300178u
Design of Bcl-2 and Bcl-xL inhibitors with subnanomolar binding affinities based upon a new scaffold
Erratum in
- J Med Chem. 2012 Jun 28;55(12):5987
Abstract
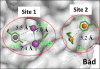
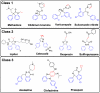
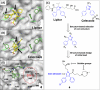
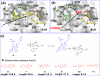
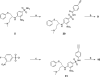
Employing a structure-based strategy, we have designed a new class of potent small-molecule inhibitors of the anti-apoptotic proteins Bcl-2 and Bcl-xL. An initial lead compound with a new scaffold was designed based upon the crystal structure of Bcl-xL and U.S. Food and Drug Administration (FDA) approved drugs and was found to have an affinity of 100 μM for both Bcl-2 and Bcl-xL. Linking this weak lead to another weak-affinity fragment derived from Abbott's ABT-737 led to an improvement of the binding affinity by a factor of >10 000. Further optimization ultimately yielded compounds with subnanomolar binding affinities for both Bcl-2 and Bcl-xL and potent cellular activity. The best compound (21) binds to Bcl-xL and Bcl-2 with K(i) < 1 nM, inhibits cell growth in the H146 and H1417 small-cell lung cancer cell lines with IC(50) values of 60-90 nM, and induces robust cell death in the H146 cancer cell line at 30-100 nM.
Figures
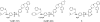

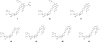

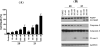
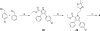
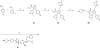
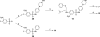
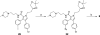
Similar articles
-
Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity.J Med Chem. 2012 Jul 12;55(13):6149-61. doi: 10.1021/jm300608w. Epub 2012 Jul 2. J Med Chem. 2012. PMID: 22747598 Free PMC article.
-
Structure-based discovery of BM-957 as a potent small-molecule inhibitor of Bcl-2 and Bcl-xL capable of achieving complete tumor regression.J Med Chem. 2012 Oct 11;55(19):8502-14. doi: 10.1021/jm3010306. Epub 2012 Oct 2. J Med Chem. 2012. PMID: 23030453 Free PMC article.
-
Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL.J Med Chem. 2007 Feb 22;50(4):641-62. doi: 10.1021/jm061152t. Epub 2007 Jan 26. J Med Chem. 2007. PMID: 17256834
-
[Progress in small-molecule inhibitors of Bcl-2 family proteins].Yao Xue Xue Bao. 2008 Jul;43(7):669-77. Yao Xue Xue Bao. 2008. PMID: 18819468 Review. Chinese.
-
Small molecule inhibition of the Bcl-X(L)-BH3 protein-protein interaction: proof-of-concept of an in vivo chemopotentiator ABT-737.Curr Top Med Chem. 2007;7(10):961-5. doi: 10.2174/156802607780906843. Curr Top Med Chem. 2007. PMID: 17508927 Review.
Cited by
-
Discovery, development and application of drugs targeting BCL-2 pro-survival proteins in cancer.Biochem Soc Trans. 2021 Nov 1;49(5):2381-2395. doi: 10.1042/BST20210749. Biochem Soc Trans. 2021. PMID: 34515749 Free PMC article. Review.
-
Inhibition of α-helix-mediated protein-protein interactions using designed molecules.Nat Chem. 2013 Mar;5(3):161-73. doi: 10.1038/nchem.1568. Nat Chem. 2013. PMID: 23422557 Review.
-
Structure-based design of potent Bcl-2/Bcl-xL inhibitors with strong in vivo antitumor activity.J Med Chem. 2012 Jul 12;55(13):6149-61. doi: 10.1021/jm300608w. Epub 2012 Jul 2. J Med Chem. 2012. PMID: 22747598 Free PMC article.
-
SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression.Cancer Res. 2014 Oct 15;74(20):5855-65. doi: 10.1158/0008-5472.CAN-14-0799. Epub 2014 Aug 21. Cancer Res. 2014. PMID: 25145672 Free PMC article.
-
TIMBAL v2: update of a database holding small molecules modulating protein-protein interactions.Database (Oxford). 2013 Jun 13;2013:bat039. doi: 10.1093/database/bat039. Print 2013. Database (Oxford). 2013. PMID: 23766369 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
-
- Ziegler DS, Kung AL. Therapeutic targeting of apoptosis pathways in cancer. Curr Opin Oncol. 2008;20(1):97–103. - PubMed
-
- Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1(2):111–121. - PubMed
-
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–656. - PubMed
-
- Cory S, Adams JM. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell. 2005;8(1):5–6. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

