Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming
- PMID: 22442037
- PMCID: PMC3334704
- DOI: 10.1261/rna.032292.112
Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming
Erratum in
- RNA. 2014 Aug;20(8):1349
Abstract
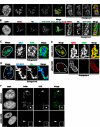
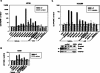
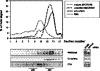
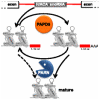
Small nucleolar and small Cajal body RNAs (snoRNAs and scaRNAs) of the H/ACA box and C/D box type are generated by exonucleolytic shortening of longer precursors. Removal of the last few nucleotides at the 3' end is known to be a distinct step. We report that, in human cells, knock-down of the poly(A) specific ribonuclease (PARN), previously implicated only in mRNA metabolism, causes the accumulation of oligoadenylated processing intermediates of H/ACA box but not C/D box RNAs. In agreement with a role of PARN in snoRNA and scaRNA processing, the enzyme is concentrated in nucleoli and Cajal bodies. Oligo(A) tails are attached to a short stub of intron sequence remaining beyond the mature 3' end of the snoRNAs. The noncanonical poly(A) polymerase PAPD5 is responsible for addition of the oligo(A) tails. We suggest that deadenylation is coupled to clean 3' end trimming, which might serve to enhance snoRNA stability.
Figures
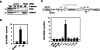


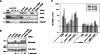
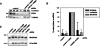
Similar articles
-
A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs.EMBO J. 2003 Aug 15;22(16):4283-93. doi: 10.1093/emboj/cdg394. EMBO J. 2003. PMID: 12912925 Free PMC article.
-
Assembly and trafficking of box C/D and H/ACA snoRNPs.RNA Biol. 2017 Jun 3;14(6):680-692. doi: 10.1080/15476286.2016.1243646. Epub 2016 Oct 7. RNA Biol. 2017. PMID: 27715451 Free PMC article. Review.
-
Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs.Nucleic Acids Res. 2007 Jan;35(Database issue):D183-7. doi: 10.1093/nar/gkl873. Epub 2006 Nov 11. Nucleic Acids Res. 2007. PMID: 17099227 Free PMC article.
-
Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus.Mol Cell Biol. 2007 Jun;27(11):4157-65. doi: 10.1128/MCB.00120-07. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403903 Free PMC article.
-
RNA modification in Cajal bodies.RNA Biol. 2017 Jun 3;14(6):693-700. doi: 10.1080/15476286.2016.1249091. Epub 2016 Oct 24. RNA Biol. 2017. PMID: 27775477 Free PMC article. Review.
Cited by
-
The PARN deadenylase targets a discrete set of mRNAs for decay and regulates cell motility in mouse myoblasts.PLoS Genet. 2012;8(8):e1002901. doi: 10.1371/journal.pgen.1002901. Epub 2012 Aug 30. PLoS Genet. 2012. PMID: 22956911 Free PMC article.
-
Deep Sequence Analysis of AgoshRNA Processing Reveals 3' A Addition and Trimming.Mol Ther Nucleic Acids. 2015 Jul 14;4(7):e247. doi: 10.1038/mtna.2015.19. Mol Ther Nucleic Acids. 2015. PMID: 26172504 Free PMC article.
-
Disruption of Telomerase RNA Maturation Kinetics Precipitates Disease.Mol Cell. 2019 May 16;74(4):688-700.e3. doi: 10.1016/j.molcel.2019.02.033. Epub 2019 Mar 28. Mol Cell. 2019. PMID: 30930056 Free PMC article.
-
Grad-seq in a Gram-positive bacterium reveals exonucleolytic sRNA activation in competence control.EMBO J. 2020 May 4;39(9):e103852. doi: 10.15252/embj.2019103852. Epub 2020 Mar 30. EMBO J. 2020. PMID: 32227509 Free PMC article.
-
Loss of the RNA helicase SKIV2L2 impairs mitotic progression and replication-dependent histone mRNA turnover in murine cell lines.RNA. 2017 Jun;23(6):910-926. doi: 10.1261/rna.060640.117. Epub 2017 Mar 28. RNA. 2017. PMID: 28351885 Free PMC article.
References
-
- Andersen JS, Lam YW, Leung AKL, Ong S-E, Lyon CE, Lamond AI, Mann M 2005. Nucleolar proteome dynamics. Nature 433: 77–83 - PubMed
-
- Bachellerie J-P, Cavaillé J, Hüttenhofer A 2002. The expanding snoRNA world. Biochimie 84: 775–790 - PubMed
-
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
