Alkbh2 protects against lethality and mutation in primary mouse embryonic fibroblasts
- PMID: 22429847
- PMCID: PMC3614354
- DOI: 10.1016/j.dnarep.2012.02.005
Alkbh2 protects against lethality and mutation in primary mouse embryonic fibroblasts
Abstract
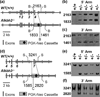
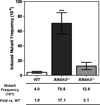
Alkylating agents modify DNA and RNA forming adducts that disrupt replication and transcription, trigger cell cycle checkpoints and/or initiate apoptosis. If left unrepaired, some of the damage can be cytotoxic and/or mutagenic. In Escherichia coli, the alkylation repair protein B (AlkB) provides one form of resistance to alkylating agents by eliminating mainly 1-methyladenine and 3-methylcytosine, thereby increasing survival and preventing mutation. To examine the biological role of the mammalian AlkB homologs Alkbh2 and Alkbh3, which both have similar enzymatic activities to that of AlkB, we evaluated the survival and mutagenesis of primary Big Blue mouse embryonic fibroblasts (MEFs) that had targeted deletions in the Alkbh2 or Alkbh3 genes. Both Alkbh2- and Alkbh3-deficient MEFs were ∼2-fold more sensitive to methyl methanesulfonate (MMS) induced cytotoxicity compared to the wild type control cells. Spontaneous mutant frequencies were similar for the wild type, Alkbh2-/- and Alkbh3-/- MEFs (average--1.3×10(-5)). However, despite the similar survival of the two mutant MEFs after MMS treatment, only the Alkbh2-deficient MEFs showed a statistically significant increase in mutant frequency compared to wild type MEFs after MMS treatment. Therefore, although both Alkbh2 and Alkbh3 can protect against MMS-induced cell death, only Alkbh2 shows statistically significant protection of MEF DNA against mutations following treatment with this exogenous methylating agent.
Copyright © 2012 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures




Similar articles
-
The DNA dioxygenase ALKBH2 protects Arabidopsis thaliana against methylation damage.Nucleic Acids Res. 2012 Aug;40(14):6620-31. doi: 10.1093/nar/gks327. Epub 2012 Apr 24. Nucleic Acids Res. 2012. PMID: 22532610 Free PMC article.
-
Lack of mismatch repair enhances resistance to methylating agents for cells deficient in oxidative demethylation.J Biol Chem. 2024 Aug;300(8):107492. doi: 10.1016/j.jbc.2024.107492. Epub 2024 Jun 24. J Biol Chem. 2024. PMID: 38925328 Free PMC article.
-
DNA repair is indispensable for survival after acute inflammation.J Clin Invest. 2012 Jul;122(7):2680-9. doi: 10.1172/JCI63338. Epub 2012 Jun 11. J Clin Invest. 2012. PMID: 22684101 Free PMC article.
-
Bacterial DNA repair genes and their eukaryotic homologues: 3. AlkB dioxygenase and Ada methyltransferase in the direct repair of alkylated DNA.Acta Biochim Pol. 2007;54(3):459-68. Epub 2007 Sep 6. Acta Biochim Pol. 2007. PMID: 17823664 Review.
-
Non-heme dioxygenases in tumor hypoxia: They're all bound with the same fate.DNA Repair (Amst). 2017 Jan;49:21-25. doi: 10.1016/j.dnarep.2016.12.001. Epub 2016 Dec 5. DNA Repair (Amst). 2017. PMID: 27964836 Review.
Cited by
-
Fluorescence Probes for ALKBH2 Allow the Measurement of DNA Alkylation Repair and Drug Resistance Responses.Angew Chem Int Ed Engl. 2018 Sep 24;57(39):12896-12900. doi: 10.1002/anie.201807593. Epub 2018 Sep 3. Angew Chem Int Ed Engl. 2018. PMID: 30098084 Free PMC article.
-
Tumor-Associated Inflammation: The Tumor-Promoting Immunity in the Early Stages of Tumorigenesis.J Immunol Res. 2022 Jun 13;2022:3128933. doi: 10.1155/2022/3128933. eCollection 2022. J Immunol Res. 2022. PMID: 35733919 Free PMC article. Review.
-
ALKBH7 drives a tissue and sex-specific necrotic cell death response following alkylation-induced damage.Cell Death Dis. 2017 Jul 20;8(7):e2947. doi: 10.1038/cddis.2017.343. Cell Death Dis. 2017. PMID: 28726787 Free PMC article.
-
Human ALKBH6 Is Required for Maintenance of Genomic Stability and Promoting Cell Survival During Exposure of Alkylating Agents in Pancreatic Cancer.Front Genet. 2021 Apr 7;12:635808. doi: 10.3389/fgene.2021.635808. eCollection 2021. Front Genet. 2021. PMID: 33897761 Free PMC article.
-
Roles of Aag, Alkbh2, and Alkbh3 in the Repair of Carboxymethylated and Ethylated Thymidine Lesions.ACS Chem Biol. 2016 May 20;11(5):1332-8. doi: 10.1021/acschembio.6b00085. Epub 2016 Mar 4. ACS Chem Biol. 2016. PMID: 26930515 Free PMC article.
References
-
- Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. - PubMed
-
- Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353:973–977. - PubMed
-
- Flashman E, Davies SL, Yeoh KK, Schofield CJ. Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem J. 2010;427:135–142. - PubMed
-
- Begley TJ, Samson LD. AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem Sci. 2003;28:2–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

