The incredible ULKs
- PMID: 22413737
- PMCID: PMC3330011
- DOI: 10.1186/1478-811X-10-7
The incredible ULKs
Abstract
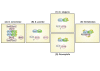
Macroautophagy (commonly abbreviated as autophagy) is an evolutionary conserved lysosome-directed vesicular trafficking pathway in eukaryotic cells that mediates the lysosomal degradation of intracellular components. The cytoplasmic cargo is initially enclosed by a specific double membrane vesicle, termed the autophagosome. By this means, autophagy either helps to remove damaged organelles, long-lived proteins and protein aggregates, or serves as a recycling mechanism for molecular building blocks. Autophagy was once invented by unicellular organisms to compensate the fluctuating external supply of nutrients. In higher eukaryotes, it is strongly enhanced under various stress conditions, such as nutrient and growth factor deprivation or DNA damage. The serine/threonine kinase Atg1 was the first identified autophagy-related gene (ATG) product in yeast. The corresponding nematode homolog UNC-51, however, has additional neuronal functions. Vertebrate genomes finally encode five closely related kinases, of which UNC-51-like kinase 1 (Ulk1) and Ulk2 are both involved in the regulation of autophagy and further neuron-specific vesicular trafficking processes. This review will mainly focus on the vertebrate Ulk1/2-Atg13-FIP200 protein complex, its function in autophagy initiation, its evolutionary descent from the yeast Atg1-Atg13-Atg17 complex, as well as the additional non-autophagic functions of its components. Since the rapid nutrient- and stress-dependent cellular responses are mainly mediated by serine/threonine phosphorylation, it will summarize our current knowledge about the relevant upstream signaling pathways and the altering phosphorylation status within this complex during autophagy induction.
Figures

Similar articles
-
A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy.Autophagy. 2009 Jul;5(5):649-62. doi: 10.4161/auto.5.5.8249. Epub 2009 Jul 20. Autophagy. 2009. PMID: 19287211
-
Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy.J Biol Chem. 2016 Mar 11;291(11):6026-6035. doi: 10.1074/jbc.M115.689646. Epub 2016 Jan 22. J Biol Chem. 2016. PMID: 26801615 Free PMC article.
-
ATG13: just a companion, or an executor of the autophagic program?Autophagy. 2014 Jun;10(6):944-56. doi: 10.4161/auto.28987. Autophagy. 2014. PMID: 24879146 Free PMC article. Review.
-
The mammalian ULK1 complex and autophagy initiation.Essays Biochem. 2017 Dec 12;61(6):585-596. doi: 10.1042/EBC20170021. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233870 Free PMC article. Review.
-
Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila.Autophagy. 2014 Mar;10(3):453-67. doi: 10.4161/auto.27442. Epub 2014 Jan 6. Autophagy. 2014. PMID: 24419107 Free PMC article.
Cited by
-
Deubiquitinase inhibition by WP1130 leads to ULK1 aggregation and blockade of autophagy.Autophagy. 2015;11(9):1458-70. doi: 10.1080/15548627.2015.1067359. Autophagy. 2015. PMID: 26207339 Free PMC article.
-
The mammalian autophagy initiator complex contains 2 HORMA domain proteins.Autophagy. 2015;11(12):2300-8. doi: 10.1080/15548627.2015.1076605. Autophagy. 2015. PMID: 26236954 Free PMC article.
-
Autophagy-independent function of Atg1 for apoptosis-induced compensatory proliferation.BMC Biol. 2016 Aug 19;14:70. doi: 10.1186/s12915-016-0293-y. BMC Biol. 2016. PMID: 27542914 Free PMC article.
-
Susceptibility of microtubule-associated protein 1 light chain 3β (MAP1LC3B/LC3B) knockout mice to lung injury and fibrosis.FASEB J. 2019 Nov;33(11):12392-12408. doi: 10.1096/fj.201900854R. Epub 2019 Aug 20. FASEB J. 2019. PMID: 31431059 Free PMC article.
-
ULK1 phosphorylates Sec23A and mediates autophagy-induced inhibition of ER-to-Golgi traffic.BMC Cell Biol. 2017 May 10;18(1):22. doi: 10.1186/s12860-017-0138-8. BMC Cell Biol. 2017. PMID: 28486929 Free PMC article.
References
-
- Klionsky DJ, Cuervo AM, Dunn WA Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3:413–416. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

