Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance
- PMID: 22409400
- PMCID: PMC3648210
- DOI: 10.1111/j.1600-0854.2012.01352.x
Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance
Abstract
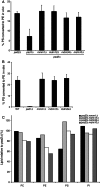
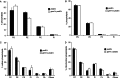
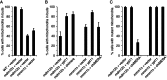
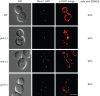
In yeast, a protein complex termed the ER-Mitochondria Encounter Structure (ERMES) tethers mitochondria to the endoplasmic reticulum. ERMES proteins are implicated in a variety of cellular functions including phospholipid synthesis, mitochondrial protein import, mitochondrial attachment to actin, polarized mitochondrial movement into daughter cells during division, and maintenance of mitochondrial DNA (mtDNA). The mitochondrial-anchored Gem1 GTPase has been proposed to regulate ERMES functions. Here, we show that ERMES and Gem1 have no direct role in the transport of phosphatidylserine (PS) from the ER to mitochondria during the synthesis of phosphatidylethanolamine (PE), as PS to PE conversion is not affected in ERMES or gem1 mutants. In addition, we report that mitochondrial inheritance defects in ERMES mutants are a secondary consequence of mitochondrial morphology defects, arguing against a primary role for ERMES in mitochondrial association with actin and mitochondrial movement. Finally, we show that ERMES complexes are long-lived, and do not depend on the presence of Gem1. Our findings suggest that the ERMES complex may have primarily a structural role in maintaining mitochondrial morphology.
© 2012 John Wiley & Sons A/S.
Figures




Similar articles
-
The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections.Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14151-6. doi: 10.1073/pnas.1111314108. Epub 2011 Aug 8. Proc Natl Acad Sci U S A. 2011. PMID: 21825164 Free PMC article.
-
Composition and topology of the endoplasmic reticulum-mitochondria encounter structure.J Mol Biol. 2011 Nov 4;413(4):743-50. doi: 10.1016/j.jmb.2011.09.012. Epub 2011 Sep 16. J Mol Biol. 2011. PMID: 21945531
-
A phospholipid transfer function of ER-mitochondria encounter structure revealed in vitro.Sci Rep. 2016 Jul 29;6:30777. doi: 10.1038/srep30777. Sci Rep. 2016. PMID: 27469264 Free PMC article.
-
The ERMES complex and ER-mitochondria connections.Biochem Soc Trans. 2012 Apr;40(2):445-50. doi: 10.1042/BST20110758. Biochem Soc Trans. 2012. PMID: 22435828 Review.
-
ER-mitochondria contact sites in yeast: beyond the myths of ERMES.Curr Opin Cell Biol. 2015 Aug;35:7-12. doi: 10.1016/j.ceb.2015.03.002. Epub 2015 Mar 30. Curr Opin Cell Biol. 2015. PMID: 25836730 Review.
Cited by
-
Lipid transport between the endoplasmic reticulum and mitochondria.Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a013235. doi: 10.1101/cshperspect.a013235. Cold Spring Harb Perspect Biol. 2013. PMID: 23732475 Free PMC article. Review.
-
Endoplasmic reticulum-mitochondria contacts: function of the junction.Nat Rev Mol Cell Biol. 2012 Oct;13(10):607-25. doi: 10.1038/nrm3440. Epub 2012 Sep 20. Nat Rev Mol Cell Biol. 2012. PMID: 22992592 Free PMC article. Review.
-
Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly.Proc Natl Acad Sci U S A. 2015 Jun 23;112(25):E3179-88. doi: 10.1073/pnas.1422363112. Epub 2015 Jun 8. Proc Natl Acad Sci U S A. 2015. PMID: 26056272 Free PMC article.
-
Hallmarks of a new era in mitochondrial biochemistry.Genes Dev. 2013 Dec 15;27(24):2615-27. doi: 10.1101/gad.229724.113. Genes Dev. 2013. PMID: 24352419 Free PMC article. Review.
-
Mitochondrial phospholipid metabolism in health and disease.J Cell Sci. 2023 Sep 1;136(17):jcs260857. doi: 10.1242/jcs.260857. Epub 2023 Sep 1. J Cell Sci. 2023. PMID: 37655851 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

