Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome
- PMID: 22403033
- PMCID: PMC3342871
- DOI: 10.1093/gbe/evs020
Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome
Abstract
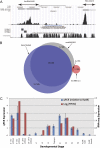
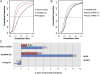
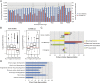
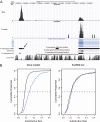
The functional repertoire of long intergenic noncoding RNA (lincRNA) molecules has begun to be elucidated in mammals. Determining the biological relevance and potential gene regulatory mechanisms of these enigmatic molecules would be expedited in a more tractable model organism, such as Drosophila melanogaster. To this end, we defined a set of 1,119 putative lincRNA genes in D. melanogaster using modENCODE whole transcriptome (RNA-seq) data. A large majority (1.1 of 1.3 Mb; 85%) of these bases were not previously reported by modENCODE as being transcribed. Significant selective constraint on the sequences of these loci predicts that virtually all have sustained functionality across the Drosophila clade. We observe biases in lincRNA genomic locations and expression profiles that are consistent with some of these lincRNAs being involved in the regulation of neighboring protein-coding genes with developmental functions. We identify lincRNAs that may be important in the developing nervous system and in male-specific organs, such as the testes. LincRNA loci were also identified whose positions, relative to nearby protein-coding loci, are equivalent between D. melanogaster and mouse. This study predicts that the genomes of not only vertebrates, such as mammals, but also an invertebrate (fruit fly) harbor large numbers of lincRNA loci. Our findings now permit exploitation of Drosophila genetics for the investigation of lincRNA mechanisms, including lincRNAs with potential functional analogues in mammals.
Figures

Similar articles
-
A Genomic Analysis of Factors Driving lincRNA Diversification: Lessons from Plants.G3 (Bethesda). 2016 Sep 8;6(9):2881-91. doi: 10.1534/g3.116.030338. G3 (Bethesda). 2016. PMID: 27440919 Free PMC article.
-
Evolutionary dynamics of lincRNA transcription in nine citrus species.Plant J. 2019 Jun;98(5):912-927. doi: 10.1111/tpj.14279. Epub 2019 Mar 18. Plant J. 2019. PMID: 30739398
-
Comparative validation of the D. melanogaster modENCODE transcriptome annotation.Genome Res. 2014 Jul;24(7):1209-23. doi: 10.1101/gr.159384.113. Genome Res. 2014. PMID: 24985915 Free PMC article.
-
Genome engineering: Drosophila melanogaster and beyond.Wiley Interdiscip Rev Dev Biol. 2016 Mar-Apr;5(2):233-67. doi: 10.1002/wdev.214. Epub 2015 Oct 8. Wiley Interdiscip Rev Dev Biol. 2016. PMID: 26447401 Free PMC article. Review.
-
Genome-wide approaches to understanding behaviour in Drosophila melanogaster.Brief Funct Genomics. 2012 Sep;11(5):395-404. doi: 10.1093/bfgp/els031. Epub 2012 Jul 26. Brief Funct Genomics. 2012. PMID: 22843979 Review.
Cited by
-
The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector.G3 (Bethesda). 2013 Sep 4;3(9):1493-509. doi: 10.1534/g3.113.006742. G3 (Bethesda). 2013. PMID: 23833213 Free PMC article.
-
Tumor suppressor miR-317 and lncRNA Peony are expressed from a polycistronic non-coding RNA locus that regulates germline differentiation and testis morphology.bioRxiv [Preprint]. 2024 Oct 10:2024.10.10.617551. doi: 10.1101/2024.10.10.617551. bioRxiv. 2024. PMID: 39416153 Free PMC article. Preprint.
-
The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription.Genes Dev. 2018 Jan 1;32(1):42-57. doi: 10.1101/gad.308619.117. Epub 2018 Jan 29. Genes Dev. 2018. PMID: 29378788 Free PMC article.
-
Transcription through enhancers suppresses their activity in Drosophila.Epigenetics Chromatin. 2013 Sep 26;6(1):31. doi: 10.1186/1756-8935-6-31. Epigenetics Chromatin. 2013. PMID: 24279291 Free PMC article.
-
Tunable self-cleaving ribozymes for modulating gene expression in eukaryotic systems.PLoS One. 2020 Apr 30;15(4):e0232046. doi: 10.1371/journal.pone.0232046. eCollection 2020. PLoS One. 2020. PMID: 32352996 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

