Polycomb repressive complex 2 is required for MLL-AF9 leukemia
- PMID: 22396593
- PMCID: PMC3324004
- DOI: 10.1073/pnas.1202258109
Polycomb repressive complex 2 is required for MLL-AF9 leukemia
Abstract
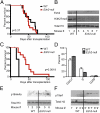
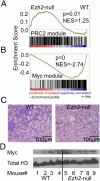
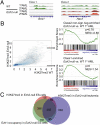
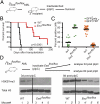
A growing body of data suggests the importance of epigenetic mechanisms in cancer. Polycomb repressive complex 2 (PRC2) has been implicated in self-renewal and cancer progression, and its components are overexpressed in many cancers. However, its role in cancer development and progression remains unclear. We used conditional alleles for the PRC2 components enhancer of zeste 2 (Ezh2) and embryonic ectoderm development (Eed) to characterize the role of PRC2 function in leukemia development and progression. Compared with wild-type leukemia, Ezh2-null MLL-AF9-mediated acute myeloid leukemia (AML) failed to accelerate upon secondary transplantation. However, Ezh2-null leukemias maintained self-renewal up to the third round of transplantation, indicating that Ezh2 is not strictly required for MLL-AF9 AML, but plays a role in leukemia progression. Genome-wide analyses of PRC2-mediated trimethylation of histone 3 demonstrated locus-specific persistence of H3K27me3 despite inactivation of Ezh2, suggesting partial compensation by Ezh1. In contrast, inactivation of the essential PRC2 gene, Eed, led to complete ablation of PRC2 function, which was incompatible with leukemia growth. Gene expression array analyses indicated more profound gene expression changes in Eed-null compared with Ezh2-null leukemic cells, including down-regulation of Myc target genes and up-regulation of PRC2 targets. Manipulating PRC2 function may be of therapeutic benefit in AML.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Inactivation of Eed impedes MLL-AF9-mediated leukemogenesis through Cdkn2a-dependent and Cdkn2a-independent mechanisms in a murine model.Exp Hematol. 2015 Nov;43(11):930-935.e6. doi: 10.1016/j.exphem.2015.06.005. Epub 2015 Jun 26. Exp Hematol. 2015. PMID: 26118502 Free PMC article.
-
Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells.Mol Cancer Ther. 2006 Dec;5(12):3096-104. doi: 10.1158/1535-7163.MCT-06-0418. Mol Cancer Ther. 2006. PMID: 17172412
-
Polycomb repressive complex 2 regulates normal development of the mouse heart.Circ Res. 2012 Feb 3;110(3):406-15. doi: 10.1161/CIRCRESAHA.111.252205. Epub 2011 Dec 8. Circ Res. 2012. PMID: 22158708 Free PMC article.
-
Inner workings and regulatory inputs that control Polycomb repressive complex 2.Chromosoma. 2012 Jun;121(3):221-34. doi: 10.1007/s00412-012-0361-1. Epub 2012 Feb 19. Chromosoma. 2012. PMID: 22349693 Free PMC article. Review.
-
EZH2 methyltransferase and H3K27 methylation in breast cancer.Int J Biol Sci. 2012;8(1):59-65. doi: 10.7150/ijbs.8.59. Epub 2011 Nov 18. Int J Biol Sci. 2012. PMID: 22211105 Free PMC article. Review.
Cited by
-
Human acute leukemia uses branched-chain amino acid catabolism to maintain stemness through regulating PRC2 function.Blood Adv. 2023 Jul 25;7(14):3592-3603. doi: 10.1182/bloodadvances.2022008242. Blood Adv. 2023. PMID: 36044390 Free PMC article.
-
Smyd2 is a Myc-regulated gene critical for MLL-AF9 induced leukemogenesis.Oncotarget. 2016 Oct 11;7(41):66398-66415. doi: 10.18632/oncotarget.12012. Oncotarget. 2016. PMID: 27655694 Free PMC article.
-
Polycomb complexes in normal and malignant hematopoiesis.J Cell Biol. 2019 Jan 7;218(1):55-69. doi: 10.1083/jcb.201808028. Epub 2018 Oct 19. J Cell Biol. 2019. PMID: 30341152 Free PMC article. Review.
-
Long Non-coding RNAs as Functional and Structural Chromatin Modulators in Acute Myeloid Leukemia.Front Oncol. 2019 Sep 11;9:899. doi: 10.3389/fonc.2019.00899. eCollection 2019. Front Oncol. 2019. PMID: 31572684 Free PMC article. Review.
-
Insights into novel emerging epigenetic drugs in myeloid malignancies.Ther Adv Hematol. 2019 Aug 6;10:2040620719866081. doi: 10.1177/2040620719866081. eCollection 2019. Ther Adv Hematol. 2019. PMID: 31431820 Free PMC article. Review.
References
-
- Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
-
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. - PubMed
-
- Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. - PubMed
-
- Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

