RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility
- PMID: 22393046
- PMCID: PMC3340185
- DOI: 10.1074/jbc.M111.337469
RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility
Abstract
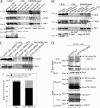
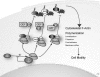
The Rho GDP dissociation inhibitor (RhoGDI) can bind to small GTPases and keep them in a biologically inactive state in cytoplasm, through which it affects actin polymerization and cell motility. However, mechanisms underlying how RhoGDI regulates Rho GTPase complex formation/membrane extraction/GTPase dissociation remain largely unexplored. Our previous studies reported that X-linked inhibitor of apoptosis protein (XIAP) interacted with RhoGDI via its RING domain and negatively modulated RhoGDI SUMOylation and HCT116 cancer cell migration. Here, we identified that RhoGDI SUMOylation specifically occurred at Lys-138, which was inhibited by XIAP domain. We further demonstrated that RhoGDI SUMOylation at Lys-138 was crucial for inhibiting actin polymerization and cytoskeleton formation as well as cancer cell motility. Moreover, SUMO-RhoGDI had a much higher binding affinity to small Rho GTPase compared with the un-SUMOylated form of RhoGDI. Taken together, our study demonstrated a novel modification of RhoGDI, SUMOylation at Lys-138, which played a key role in regulating Rho GTPase activation in cancer cells. The physiological regulation of RhoGDI SUMOylation by the RING domain of XIAP may account for modulation of cancer cell invasion and metastasis by XIAP.
Figures


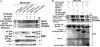
Similar articles
-
X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton.J Biol Chem. 2011 May 6;286(18):15630-40. doi: 10.1074/jbc.M110.176982. Epub 2011 Mar 14. J Biol Chem. 2011. PMID: 21402697 Free PMC article.
-
E3 ligase activity of XIAP RING domain is required for XIAP-mediated cancer cell migration, but not for its RhoGDI binding activity.PLoS One. 2012;7(4):e35682. doi: 10.1371/journal.pone.0035682. Epub 2012 Apr 19. PLoS One. 2012. PMID: 22532870 Free PMC article.
-
ExoS Rho GTPase-activating protein activity stimulates reorganization of the actin cytoskeleton through Rho GTPase guanine nucleotide disassociation inhibitor.J Biol Chem. 2004 Oct 8;279(41):42936-44. doi: 10.1074/jbc.M406493200. Epub 2004 Aug 2. J Biol Chem. 2004. PMID: 15292224
-
The 'invisible hand': regulation of RHO GTPases by RHOGDIs.Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):493-504. doi: 10.1038/nrm3153. Nat Rev Mol Cell Biol. 2011. PMID: 21779026 Free PMC article. Review.
-
RhoGDI: multiple functions in the regulation of Rho family GTPase activities.Biochem J. 2005 Aug 15;390(Pt 1):1-9. doi: 10.1042/BJ20050104. Biochem J. 2005. PMID: 16083425 Free PMC article. Review.
Cited by
-
MicroRNA-3648 Is Upregulated to Suppress TCF21, Resulting in Promotion of Invasion and Metastasis of Human Bladder Cancer.Mol Ther Nucleic Acids. 2019 Jun 7;16:519-530. doi: 10.1016/j.omtn.2019.04.006. Epub 2019 Apr 14. Mol Ther Nucleic Acids. 2019. PMID: 31071528 Free PMC article.
-
Comprehensive identification of SUMO2/3 targets and their dynamics during mitosis.PLoS One. 2014 Jun 27;9(6):e100692. doi: 10.1371/journal.pone.0100692. eCollection 2014. PLoS One. 2014. PMID: 24971888 Free PMC article.
-
High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression.Oncotarget. 2014 Sep 15;5(17):7458-70. doi: 10.18632/oncotarget.2150. Oncotarget. 2014. PMID: 25277185 Free PMC article.
-
Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.Nat Chem Biol. 2017 Sep;13(9):994-1001. doi: 10.1038/nchembio.2442. Epub 2017 Jul 24. Nat Chem Biol. 2017. PMID: 28759015
-
X-linked inhibitor of apoptosis protein (XIAP) regulation of cyclin D1 protein expression and cancer cell anchorage-independent growth via its E3 ligase-mediated protein phosphatase 2A/c-Jun axis.J Biol Chem. 2013 Jul 12;288(28):20238-47. doi: 10.1074/jbc.M112.448365. Epub 2013 May 29. J Biol Chem. 2013. PMID: 23720779 Free PMC article.
References
-
- Kaibuchi K., Kuroda S., Amano M. (1999) Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 68, 459–486 - PubMed
-
- Olofsson B. (1999) Rho guanine dissociation inhibitors. Pivotal molecules in cellular signaling. Cell. Signal. 11, 545–554 - PubMed
-
- Etienne-Manneville S., Hall A. (2002) Rho GTPases in cell biology. Nature 420, 629–635 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

