Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease
- PMID: 22388089
- PMCID: PMC3364717
- DOI: 10.1038/nm.2667
Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease
Abstract
During chronic injury a population of bipotent hepatic progenitor cells (HPCs) become activated to regenerate both cholangiocytes and hepatocytes. Here we show in human diseased liver and mouse models of the ductular reaction that Notch and Wnt signaling direct specification of HPCs via their interactions with activated myofibroblasts or macrophages. In particular, we found that during biliary regeneration, expression of Jagged 1 (a Notch ligand) by myofibroblasts promoted Notch signaling in HPCs and thus their biliary specification to cholangiocytes. Alternatively, during hepatocyte regeneration, macrophage engulfment of hepatocyte debris induced Wnt3a expression. This resulted in canonical Wnt signaling in nearby HPCs, thus maintaining expression of Numb (a cell fate determinant) within these cells and the promotion of their specification to hepatocytes. By these two pathways adult parenchymal regeneration during chronic liver injury is promoted.
Figures
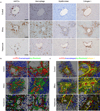
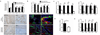
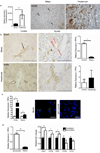

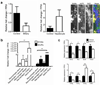
Comment in
-
Neighborhood watch orchestrates liver regeneration.Nat Med. 2012 Apr 5;18(4):497-9. doi: 10.1038/nm.2719. Nat Med. 2012. PMID: 22481408 No abstract available.
-
The balance between Notch/Wnt signaling regulates progenitor cells' commitment during liver repair: mystery solved?J Hepatol. 2013 Jan;58(1):181-3. doi: 10.1016/j.jhep.2012.08.006. Epub 2012 Aug 15. J Hepatol. 2013. PMID: 22902547 No abstract available.
-
The role of paracrine signals during liver regeneration.Hepatology. 2012 Oct;56(4):1577-9. doi: 10.1002/hep.25911. Hepatology. 2012. PMID: 23038651 Free PMC article.
Similar articles
-
Enhanced Wnt/β-catenin and Notch signalling in the activated canine hepatic progenitor cell niche.BMC Vet Res. 2014 Dec 31;10:309. doi: 10.1186/s12917-014-0309-1. BMC Vet Res. 2014. PMID: 25551829 Free PMC article.
-
Notch Signaling Coordinates Progenitor Cell-Mediated Biliary Regeneration Following Partial Hepatectomy.Sci Rep. 2016 Mar 8;6:22754. doi: 10.1038/srep22754. Sci Rep. 2016. PMID: 26951801 Free PMC article.
-
The wnt target jagged-1 mediates the activation of notch signaling by progastrin in human colorectal cancer cells.Cancer Res. 2009 Aug 1;69(15):6065-73. doi: 10.1158/0008-5472.CAN-08-2409. Epub 2009 Jul 21. Cancer Res. 2009. PMID: 19622776
-
Notch signalling beyond liver development: emerging concepts in liver repair and oncogenesis.Clin Res Hepatol Gastroenterol. 2013 Nov;37(5):447-54. doi: 10.1016/j.clinre.2013.05.008. Epub 2013 Jun 24. Clin Res Hepatol Gastroenterol. 2013. PMID: 23806629 Review.
-
Cell and molecular biology of Notch.J Endocrinol. 2007 Sep;194(3):459-74. doi: 10.1677/JOE-07-0242. J Endocrinol. 2007. PMID: 17761886 Review.
Cited by
-
Notch signaling in hepatocellular carcinoma: guilty in association!Gastroenterology. 2012 Dec;143(6):1430-4. doi: 10.1053/j.gastro.2012.10.025. Epub 2012 Oct 22. Gastroenterology. 2012. PMID: 23099244 Free PMC article. No abstract available.
-
FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration.Genes Dev. 2013 Jan 15;27(2):169-81. doi: 10.1101/gad.204776.112. Epub 2013 Jan 15. Genes Dev. 2013. PMID: 23322300 Free PMC article.
-
Kupffer cells induce Notch-mediated hepatocyte conversion in a common mouse model of intrahepatic cholangiocarcinoma.Sci Rep. 2016 Oct 4;6:34691. doi: 10.1038/srep34691. Sci Rep. 2016. PMID: 27698452 Free PMC article.
-
Epistatic Association of CD14 and NOTCH2 Genetic Polymorphisms with Biliary Atresia in a Southern Chinese Population.Mol Ther Nucleic Acids. 2018 Dec 7;13:590-595. doi: 10.1016/j.omtn.2018.10.006. Epub 2018 Oct 16. Mol Ther Nucleic Acids. 2018. PMID: 30439647 Free PMC article.
-
M2 macrophages promote myofibroblast differentiation of LR-MSCs and are associated with pulmonary fibrogenesis.Cell Commun Signal. 2018 Nov 23;16(1):89. doi: 10.1186/s12964-018-0300-8. Cell Commun Signal. 2018. PMID: 30470231 Free PMC article.
References
-
- World Health Organisation Disease and injury country estimates, Burden of disease. 2009 http://www.who.int/healthinfo/global_burden_disease/estimates_country/en.... Ref Type: Generic.
-
- Hay DC. Cadaveric hepatocytes repopulate diseased livers: life after death. Gastroenterology. 2010;139:729–731. - PubMed
-
- Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2010 - PubMed
-
- Fellous TG, et al. Locating the stem cell niche and tracing hepatocyte lineages in human liver. Hepatology. 2009;49:1655–1663. - PubMed
-
- Gouw AS, Clouston AD, Theise ND. Ductular reactions in human liver: Diversity at the interface. Hepatology. 2011;54:1853–1863. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

