Genetic diversity of EBV-encoded LMP1 in the Swiss HIV Cohort Study and implication for NF-Κb activation
- PMID: 22384168
- PMCID: PMC3285206
- DOI: 10.1371/journal.pone.0032168
Genetic diversity of EBV-encoded LMP1 in the Swiss HIV Cohort Study and implication for NF-Κb activation
Abstract
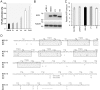
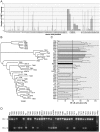
Epstein-Barr virus (EBV) is associated with several types of cancers including Hodgkin's lymphoma (HL) and nasopharyngeal carcinoma (NPC). EBV-encoded latent membrane protein 1 (LMP1), a multifunctional oncoprotein, is a powerful activator of the transcription factor NF-κB, a property that is essential for EBV-transformed lymphoblastoid cell survival. Previous studies reported LMP1 sequence variations and induction of higher NF-κB activation levels compared to the prototype B95-8 LMP1 by some variants. Here we used biopsies of EBV-associated cancers and blood of individuals included in the Swiss HIV Cohort Study (SHCS) to analyze LMP1 genetic diversity and impact of sequence variations on LMP1-mediated NF-κB activation potential. We found that a number of variants mediate higher NF-κB activation levels when compared to B95-8 LMP1 and mapped three single polymorphisms responsible for this phenotype: F106Y, I124V and F144I. F106Y was present in all LMP1 isolated in this study and its effect was variant dependent, suggesting that it was modulated by other polymorphisms. The two polymorphisms I124V and F144I were present in distinct phylogenetic groups and were linked with other specific polymorphisms nearby, I152L and D150A/L151I, respectively. The two sets of polymorphisms, I124V/I152L and F144I/D150A/L151I, which were markers of increased NF-κB activation in vitro, were not associated with EBV-associated HL in the SHCS. Taken together these results highlighted the importance of single polymorphisms for the modulation of LMP1 signaling activity and demonstrated that several groups of LMP1 variants, through distinct mutational paths, mediated enhanced NF-κB activation levels compared to B95-8 LMP1.
Conflict of interest statement
Figures

Similar articles
-
[Functional analysis of Epstein-Barr virus latent membrane proteins (LMP1) in patients with limphoproliferative disorders].Biomed Khim. 2011 Jan-Feb;57(1):114-26. doi: 10.18097/pbmc20115701114. Biomed Khim. 2011. PMID: 21516783 Russian.
-
Nucleotide sequences and functions of the Epstein-Barr virus latent membrane protein 1 genes isolated from salivary gland lymphoepithelial carcinomas.Virus Genes. 2005 Mar;30(2):223-35. doi: 10.1007/s11262-004-5630-5. Virus Genes. 2005. PMID: 15744579
-
The tumor marker Fascin is induced by the Epstein-Barr virus-encoded oncoprotein LMP1 via NF-κB in lymphocytes and contributes to their invasive migration.Cell Commun Signal. 2014 Jul 11;12:46. doi: 10.1186/s12964-014-0046-x. Cell Commun Signal. 2014. PMID: 25105941 Free PMC article.
-
Epstein-barr virus transformation: involvement of latent membrane protein 1-mediated activation of NF-kappaB.Oncogene. 1999 Nov 22;18(49):6959-64. doi: 10.1038/sj.onc.1203217. Oncogene. 1999. PMID: 10602470 Review.
-
NF-κB and IRF7 pathway activation by Epstein-Barr virus Latent Membrane Protein 1.Viruses. 2013 Jun 21;5(6):1587-606. doi: 10.3390/v5061587. Viruses. 2013. PMID: 23793113 Free PMC article. Review.
Cited by
-
Evaluation of latent membrane protein 1 and microRNA-155 for the prognostic prediction of diffuse large B cell lymphoma.Oncol Lett. 2018 Jun;15(6):9725-9734. doi: 10.3892/ol.2018.8560. Epub 2018 Apr 24. Oncol Lett. 2018. PMID: 29844839 Free PMC article.
-
Genomic Instability: The Driving Force behind Refractory/Relapsing Hodgkin's Lymphoma.Cancers (Basel). 2013 Jun 5;5(2):714-25. doi: 10.3390/cancers5020714. Cancers (Basel). 2013. PMID: 24216998 Free PMC article.
-
Epstein-barr virus sequence variation-biology and disease.Pathogens. 2012 Nov 8;1(2):156-74. doi: 10.3390/pathogens1020156. Pathogens. 2012. PMID: 25436768 Free PMC article. Review.
-
A novel recombinant variant of latent membrane protein 1 from Epstein Barr virus in Argentina denotes phylogeographical association.PLoS One. 2017 Mar 22;12(3):e0174221. doi: 10.1371/journal.pone.0174221. eCollection 2017. PLoS One. 2017. PMID: 28328987 Free PMC article.
-
Molecular characterization of Epstein-Barr virus variants detected in the oral cavity of adolescents in Cali, Colombia.Biomedica. 2020 May 1;40(Supl. 1):76-88. doi: 10.7705/biomedica.4917. Biomedica. 2020. PMID: 32463610 Free PMC article. English, Spanish.
References
-
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. - PubMed
-
- Dirmeier U, Neuhierl B, Kilger E, Reisbach G, Sandberg ML, et al. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by epstein-barr virus. Cancer Res. 2003;63:2982–2989. - PubMed
-
- Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

