West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response
- PMID: 22379089
- PMCID: PMC3347305
- DOI: 10.1128/JVI.00207-12
West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response
Abstract
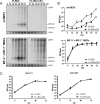
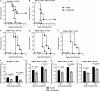
We previously showed that a noncoding subgenomic flavivirus RNA (sfRNA) is required for viral pathogenicity, as a mutant West Nile virus (WNV) deficient in sfRNA production replicated poorly in wild-type mice. To investigate the possible immunomodulatory or immune evasive functions of sfRNA, we utilized mice and cells deficient in elements of the type I interferon (IFN) response. Replication of the sfRNA mutant WNV was rescued in mice and cells lacking interferon regulatory factor 3 (IRF-3) and IRF-7 and in mice lacking the type I alpha/beta interferon receptor (IFNAR), suggesting a contribution for sfRNA in overcoming the antiviral response mediated by type I IFN. This was confirmed by demonstrating rescue of mutant virus replication in the presence of IFNAR neutralizing antibodies, greater sensitivity of mutant virus replication to IFN-α pretreatment, partial rescue of its infectivity in cells deficient in RNase L, and direct effects of transfected sfRNA on rescuing replication of unrelated Semliki Forest virus in cells pretreated with IFN-α. The results define a novel function of sfRNA in flavivirus pathogenesis via its contribution to viral evasion of the type I interferon response.
Figures

Similar articles
-
Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes.J Virol. 2016 Oct 28;90(22):10145-10159. doi: 10.1128/JVI.00930-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581979 Free PMC article.
-
Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses.Viruses. 2014 Jan 27;6(2):404-27. doi: 10.3390/v6020404. Viruses. 2014. PMID: 24473339 Free PMC article. Review.
-
A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice.J Virol. 2006 Mar;80(5):2396-404. doi: 10.1128/JVI.80.5.2396-2404.2006. J Virol. 2006. PMID: 16474146 Free PMC article.
-
Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection.J Virol. 2008 Sep;82(17):8465-75. doi: 10.1128/JVI.00918-08. Epub 2008 Jun 18. J Virol. 2008. PMID: 18562536 Free PMC article.
-
Functional non-coding RNAs derived from the flavivirus 3' untranslated region.Virus Res. 2015 Aug 3;206:53-61. doi: 10.1016/j.virusres.2015.01.026. Epub 2015 Feb 7. Virus Res. 2015. PMID: 25660582 Review.
Cited by
-
Rasputin a decade on and more promiscuous than ever? A review of G3BPs.Biochim Biophys Acta Mol Cell Res. 2019 Mar;1866(3):360-370. doi: 10.1016/j.bbamcr.2018.09.001. Epub 2018 Sep 5. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30595162 Free PMC article. Review.
-
JEV Infection Induces M-MDSC Differentiation Into CD3+ Macrophages in the Brain.Front Immunol. 2022 Apr 21;13:838990. doi: 10.3389/fimmu.2022.838990. eCollection 2022. Front Immunol. 2022. PMID: 35529855 Free PMC article.
-
Non-coding RNA function in stem cells and Regenerative Medicine.Noncoding RNA Res. 2018 Apr 20;3(2):39-41. doi: 10.1016/j.ncrna.2018.04.004. eCollection 2018 Jun. Noncoding RNA Res. 2018. PMID: 30159438 Free PMC article. No abstract available.
-
A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.Proc Natl Acad Sci U S A. 2018 Jun 19;115(25):6404-6409. doi: 10.1073/pnas.1802429115. Epub 2018 Jun 4. Proc Natl Acad Sci U S A. 2018. PMID: 29866852 Free PMC article.
-
Chikungunya virus 3' untranslated region: adaptation to mosquitoes and a population bottleneck as major evolutionary forces.PLoS Pathog. 2013;9(8):e1003591. doi: 10.1371/journal.ppat.1003591. Epub 2013 Aug 29. PLoS Pathog. 2013. PMID: 24009512 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

