Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells
- PMID: 22375014
- PMCID: PMC3339927
- DOI: 10.1074/jbc.M111.333963
Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells
Abstract
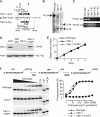
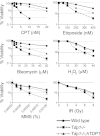
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) repairs topoisomerase I cleavage complexes (Top1cc) by hydrolyzing their 3'-phosphotyrosyl DNA bonds and repairs bleomycin-induced DNA damage by hydrolyzing 3'-phosphoglycolates. Yeast Tdp1 has also been implicated in the repair of topoisomerase II-DNA cleavage complexes (Top2cc). To determine whether vertebrate Tdp1 is involved in the repair of various DNA end-blocking lesions, we generated Tdp1 knock-out cells in chicken DT40 cells (Tdp1-/-) and Tdp1-complemented DT40 cells with human TDP1. We found that Tdp1-/- cells were not only hypersensitive to camptothecin and bleomycin but also to etoposide, methyl methanesulfonate (MMS), H(2)O(2), and ionizing radiation. We also show they were deficient in mitochondrial Tdp1 activity. In biochemical assays, recombinant human TDP1 was found to process 5'-phosphotyrosyl DNA ends when they mimic the 5'-overhangs of Top2cc. Tdp1 also processes 3'-deoxyribose phosphates generated from hydrolysis of abasic sites, which is consistent with the hypersensitivity of Tdp1-/- cells to MMS and H(2)O(2). Because recent studies established that CtIP together with BRCA1 also repairs topoisomerase-mediated DNA damage, we generated dual Tdp1-CtIP-deficient DT40 cells. Our results show that Tdp1 and CtIP act in parallel pathways for the repair of Top1cc and MMS-induced lesions but are epistatic for Top2cc. Together, our findings reveal a broad involvement of Tdp1 in DNA repair and clarify the role of human TDP1 in the repair of Top2-induced DNA damage.
Figures





Similar articles
-
Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage.Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):8953-8. doi: 10.1073/pnas.0603455103. Epub 2006 Jun 2. Proc Natl Acad Sci U S A. 2006. PMID: 16751265 Free PMC article.
-
Tyrosyl-DNA-phosphodiesterase I (TDP1) participates in the removal and repair of stabilized-Top2α cleavage complexes in human cells.Mutat Res. 2015 Nov;781:37-48. doi: 10.1016/j.mrfmmm.2015.09.003. Epub 2015 Sep 14. Mutat Res. 2015. PMID: 26421495
-
PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage.Nucleic Acids Res. 2014 Apr;42(7):4435-49. doi: 10.1093/nar/gku088. Epub 2014 Feb 3. Nucleic Acids Res. 2014. PMID: 24493735 Free PMC article.
-
Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2).DNA Repair (Amst). 2014 Jul;19:114-29. doi: 10.1016/j.dnarep.2014.03.020. Epub 2014 May 22. DNA Repair (Amst). 2014. PMID: 24856239 Free PMC article. Review.
-
Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation.Nucleic Acids Res. 2018 Jan 25;46(2):520-537. doi: 10.1093/nar/gkx1219. Nucleic Acids Res. 2018. PMID: 29216365 Free PMC article. Review.
Cited by
-
Neurological disorders associated with DNA strand-break processing enzymes.Mech Ageing Dev. 2017 Jan;161(Pt A):130-140. doi: 10.1016/j.mad.2016.07.009. Epub 2016 Jul 25. Mech Ageing Dev. 2017. PMID: 27470939 Free PMC article. Review.
-
Participation of TDP1 in the repair of formaldehyde-induced DNA-protein cross-links in chicken DT40 cells.PLoS One. 2020 Jun 26;15(6):e0234859. doi: 10.1371/journal.pone.0234859. eCollection 2020. PLoS One. 2020. PMID: 32589683 Free PMC article.
-
Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways.DNA Repair (Amst). 2020 Oct;94:102926. doi: 10.1016/j.dnarep.2020.102926. Epub 2020 Jul 10. DNA Repair (Amst). 2020. PMID: 32674013 Free PMC article. Review.
-
Novel Tdp1 Inhibitors Based on Adamantane Connected with Monoterpene Moieties via Heterocyclic Fragments.Molecules. 2021 May 24;26(11):3128. doi: 10.3390/molecules26113128. Molecules. 2021. PMID: 34073771 Free PMC article.
-
Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes.J Biol Chem. 2012 Aug 31;287(36):30842-52. doi: 10.1074/jbc.M112.393983. Epub 2012 Jul 20. J Biol Chem. 2012. PMID: 22822062 Free PMC article.
References
-
- Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 - PubMed
-
- Stewart L., Redinbo M. R., Qiu X., Hol W. G., Champoux J. J. (1998) A model for the mechanism of human topoisomerase I. Science 279, 1534–1541 - PubMed
-
- Koster D. A., Croquette V., Dekker C., Shuman S., Dekker N. H. (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

