Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography
- PMID: 22371572
- PMCID: PMC3306676
- DOI: 10.1073/pnas.1120453109
Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography
Abstract
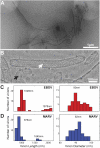
Ebola virus is a highly pathogenic filovirus causing severe hemorrhagic fever with high mortality rates. It assembles heterogenous, filamentous, enveloped virus particles containing a negative-sense, single-stranded RNA genome packaged within a helical nucleocapsid (NC). We have used cryo-electron microscopy and tomography to visualize Ebola virus particles, as well as Ebola virus-like particles, in three dimensions in a near-native state. The NC within the virion forms a left-handed helix with an inner nucleoprotein layer decorated with protruding arms composed of VP24 and VP35. A comparison with the closely related Marburg virus shows that the N-terminal region of nucleoprotein defines the inner diameter of the Ebola virus NC, whereas the RNA genome defines its length. Binding of the nucleoprotein to RNA can assemble a loosely coiled NC-like structure; the loose coil can be condensed by binding of the viral matrix protein VP40 to the C terminus of the nucleoprotein, and rigidified by binding of VP24 and VP35 to alternate copies of the nucleoprotein. Four proteins (NP, VP24, VP35, and VP40) are necessary and sufficient to mediate assembly of an NC with structure, symmetry, variability, and flexibility indistinguishable from that in Ebola virus particles released from infected cells. Together these data provide a structural and architectural description of Ebola virus and define the roles of viral proteins in its structure and assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.Cell. 2024 Oct 3;187(20):5587-5603.e19. doi: 10.1016/j.cell.2024.08.044. Epub 2024 Sep 17. Cell. 2024. PMID: 39293445
-
Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 Å resolution.Nature. 2018 Nov;563(7729):137-140. doi: 10.1038/s41586-018-0630-0. Epub 2018 Oct 17. Nature. 2018. PMID: 30333622
-
The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy.PLoS One. 2012;7(1):e29608. doi: 10.1371/journal.pone.0029608. Epub 2012 Jan 11. PLoS One. 2012. PMID: 22247782 Free PMC article.
-
Filovirus helical nucleocapsid structures.Microscopy (Oxf). 2023 Jun 8;72(3):178-190. doi: 10.1093/jmicro/dfac049. Microscopy (Oxf). 2023. PMID: 36242583 Review.
-
Filovirus Filament Proteins.Subcell Biochem. 2018;88:73-94. doi: 10.1007/978-981-10-8456-0_4. Subcell Biochem. 2018. PMID: 29900493 Review.
Cited by
-
An Ebola Virus-Like Particle-Based Reporter System Enables Evaluation of Antiviral Drugs In Vivo under Non-Biosafety Level 4 Conditions.J Virol. 2016 Sep 12;90(19):8720-8. doi: 10.1128/JVI.01239-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27440895 Free PMC article.
-
An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.Cell Rep. 2015 Apr 21;11(3):376-89. doi: 10.1016/j.celrep.2015.03.034. Epub 2015 Apr 9. Cell Rep. 2015. PMID: 25865894 Free PMC article.
-
The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells.Sci Rep. 2013;3:1206. doi: 10.1038/srep01206. Epub 2013 Feb 4. Sci Rep. 2013. PMID: 23383374 Free PMC article.
-
Therapeutic strategies to target the Ebola virus life cycle.Nat Rev Microbiol. 2019 Oct;17(10):593-606. doi: 10.1038/s41579-019-0233-2. Epub 2019 Jul 24. Nat Rev Microbiol. 2019. PMID: 31341272 Review.
-
TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction.PLoS Pathog. 2022 May 9;18(5):e1010530. doi: 10.1371/journal.ppat.1010530. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35533151 Free PMC article.
References
-
- Sanchez A, Geisbert T, Feldmann H. Filoviridae: Marburg and Ebola viruses. In: Knipe D, Howley P, editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams and Wilkins; 2007. p. 1409.
-
- Lamb R. Mononegavirales. In: Knipe D, Howley P, editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams and Wilkins; 2007. p. 1357.
-
- Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14:504–510. - PubMed
-
- Becker S, Rinne C, Hofsäss U, Klenk H-D, Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

