Global profiling of DNA methylation erasure in mouse primordial germ cells
- PMID: 22357612
- PMCID: PMC3317146
- DOI: 10.1101/gr.130997.111
Global profiling of DNA methylation erasure in mouse primordial germ cells
Abstract
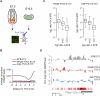
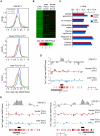
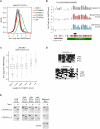
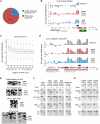
Epigenetic reprogramming, characterized by loss of cytosine methylation and histone modifications, occurs during mammalian development in primordial germ cells (PGCs), yet the targets and kinetics of this process are poorly characterized. Here we provide a map of cytosine methylation on a large portion of the genome in developing male and female PGCs isolated from mouse embryos. We show that DNA methylation erasure is global and affects genes of various biological functions. We also reveal complex kinetics of demethylation that are initiated at most genes in early PGC precursors around embryonic day 8.0-9.0. In addition, besides intracisternal A-particles (IAPs), we identify rare LTR-ERV1 retroelements and single-copy sequences that resist global methylation erasure in PGCs as well as in preimplantation embryos. Our data provide important insights into the targets and dynamics of DNA methylation reprogramming in mammalian germ cells.
Figures

Similar articles
-
Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency.Nature. 2010 Feb 25;463(7284):1101-5. doi: 10.1038/nature08829. Nature. 2010. PMID: 20098412 Free PMC article.
-
Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine.Science. 2013 Jan 25;339(6118):448-52. doi: 10.1126/science.1229277. Epub 2012 Dec 6. Science. 2013. PMID: 23223451 Free PMC article.
-
Effects of dppa3 on DNA methylation dynamics during primordial germ cell development in mice.Biol Reprod. 2013 May 23;88(5):125. doi: 10.1095/biolreprod.112.105932. Print 2013 May. Biol Reprod. 2013. PMID: 23595900
-
Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110330. doi: 10.1098/rstb.2011.0330. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166394 Free PMC article. Review.
-
Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells.Curr Opin Cell Biol. 2013 Jun;25(3):281-8. doi: 10.1016/j.ceb.2013.02.013. Epub 2013 Mar 17. Curr Opin Cell Biol. 2013. PMID: 23510682 Review.
Cited by
-
Multifaceted paternal exposures before conception and their epigenetic impact on offspring.J Assist Reprod Genet. 2024 Nov;41(11):2931-2951. doi: 10.1007/s10815-024-03243-1. Epub 2024 Sep 4. J Assist Reprod Genet. 2024. PMID: 39230664 Review.
-
Specification and epigenetic programming of the human germ line.Nat Rev Genet. 2016 Oct;17(10):585-600. doi: 10.1038/nrg.2016.88. Epub 2016 Aug 30. Nat Rev Genet. 2016. PMID: 27573372 Review.
-
The third barrier to transgenerational inheritance in animals: somatic epigenetic resetting.EMBO Rep. 2023 Apr 5;24(4):e56615. doi: 10.15252/embr.202256615. Epub 2023 Mar 2. EMBO Rep. 2023. PMID: 36862326 Free PMC article.
-
Sex-specific chromatin remodelling safeguards transcription in germ cells.Nature. 2021 Dec;600(7890):737-742. doi: 10.1038/s41586-021-04208-5. Epub 2021 Dec 8. Nature. 2021. PMID: 34880491
-
DevMouse, the mouse developmental methylome database and analysis tools.Database (Oxford). 2014 Jan 9;2014:bat084. doi: 10.1093/database/bat084. Print 2014. Database (Oxford). 2014. PMID: 24408217 Free PMC article.
References
-
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42: 1093–1100 - PubMed
-
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424 - PubMed
-
- Guibert S, Forne T, Weber M 2009. Dynamic regulation of DNA methylation during mammalian development. Epigenomics 1: 81–98 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
