A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA
- PMID: 22355643
- PMCID: PMC3216607
- DOI: 10.1038/srep00126
A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA
Abstract
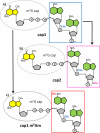
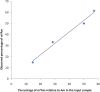
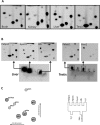
A method is described for the detection of certain nucleotide modifications adjacent to the 5' 7-methyl guanosine cap of mRNAs from individual genes. The method quantitatively measures the relative abundance of 2'-O-methyl and N(6),2'-O-dimethyladenosine, two of the most common modifications. In order to identify and quantitatify the amounts of N(6),2'-O-dimethyladenosine, a novel method for the synthesis of modified adenosine phosphoramidites was developed. This method is a one step synthesis and the product can directly be used for the production of N(6),2'-O-dimethyladenosine containing RNA oligonucleotides. The nature of the cap-adjacent nucleotides were shown to be characteristic for mRNAs from individual genes transcribed in liver and testis.
Figures

Similar articles
-
A novel approach to solid phase chemical synthesis of oligonucleotide mRNA cap analogs.Nucleosides Nucleotides Nucleic Acids. 2005;24(5-7):601-5. doi: 10.1081/ncn-200061922. Nucleosides Nucleotides Nucleic Acids. 2005. PMID: 16247996
-
Preparation of Functional, Fluorescently Labeled mRNA Capped with Anthraniloyl-m(7)GpppG.Methods Mol Biol. 2016;1428:61-75. doi: 10.1007/978-1-4939-3625-0_4. Methods Mol Biol. 2016. PMID: 27236792
-
Cap analogs containing 6-thioguanosine--reagents for the synthesis of mRNAs selectively photo-crosslinkable with cap-binding biomolecules.Org Biomol Chem. 2014 Jul 21;12(27):4841-7. doi: 10.1039/c4ob00059e. Org Biomol Chem. 2014. PMID: 24763507
-
Synthetic mRNAs with superior translation and stability properties.Methods Mol Biol. 2013;969:55-72. doi: 10.1007/978-1-62703-260-5_4. Methods Mol Biol. 2013. PMID: 23296927 Review.
-
Discovering and Mapping the Modified Nucleotides That Comprise the Epitranscriptome of mRNA.Cold Spring Harb Perspect Biol. 2019 Jun 3;11(6):a032201. doi: 10.1101/cshperspect.a032201. Cold Spring Harb Perspect Biol. 2019. PMID: 31160350 Free PMC article. Review.
Cited by
-
Quick Access to Nucleobase-Modified Phosphoramidites for the Synthesis of Oligoribonucleotides Containing Post-Transcriptional Modifications and Epitranscriptomic Marks.J Org Chem. 2022 Aug 5;87(15):10333-10348. doi: 10.1021/acs.joc.2c01390. Epub 2022 Jul 20. J Org Chem. 2022. PMID: 35857285 Free PMC article.
-
Combining Chemical Synthesis and Enzymatic Methylation to Access Short RNAs with Various 5' Caps.Chembiochem. 2019 Jul 1;20(13):1693-1700. doi: 10.1002/cbic.201900037. Epub 2019 May 27. Chembiochem. 2019. PMID: 30768827 Free PMC article.
-
m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome.Nat Methods. 2016 Aug;13(8):692-8. doi: 10.1038/nmeth.3898. Epub 2016 Jul 4. Nat Methods. 2016. PMID: 27376769 Free PMC article.
-
CMTR1-Catalyzed 2'-O-Ribose Methylation Controls Neuronal Development by Regulating Camk2α Expression Independent of RIG-I Signaling.Cell Rep. 2020 Oct 20;33(3):108269. doi: 10.1016/j.celrep.2020.108269. Cell Rep. 2020. PMID: 33086056 Free PMC article.
-
5' UTR m(6)A Promotes Cap-Independent Translation.Cell. 2015 Nov 5;163(4):999-1010. doi: 10.1016/j.cell.2015.10.012. Epub 2015 Oct 22. Cell. 2015. PMID: 26593424 Free PMC article.
References
-
- Langberg S. R. and Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2'-)-methyltransferases from HeLa cells. J. Biol. Chem., 256, 54–60 (1981). - PubMed
-
- Wei C. M., Gershowitz A. and Moss B. N6,O2-dimethyladenosine a novel ribonucleoside next to the 5'terminal of animal cell and virus mRNAs. Nature, 257, 251–253 (1975). - PubMed
-
- Keith J. M., Ensinger M. J. and Moss B. HeLa cell RNA(2'-O-methyladenosine –N6-)-methyltransferase specific for the capped 5'-end of messenger RNA. J. Biol. Chem., 253, 5033–5041 (1978). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

