Interplay between NS3 protease and human La protein regulates translation-replication switch of Hepatitis C virus
- PMID: 22355520
- PMCID: PMC3210691
- DOI: 10.1038/srep00001
Interplay between NS3 protease and human La protein regulates translation-replication switch of Hepatitis C virus
Abstract
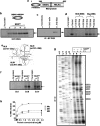
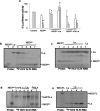
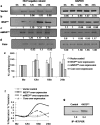
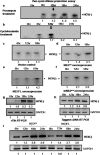
HCV NS3 protein plays a central role in viral polyprotein processing and RNA replication. We demonstrate that the NS3 protease (NS3(pro)) domain alone can specifically bind to HCV-IRES RNA, predominantly in the SLIV region. The cleavage activity of the NS3 protease domain is reduced upon HCV-RNA binding. More importantly, NS3(pro) binding to the SLIV hinders the interaction of La protein, a cellular IRES-trans acting factor required for HCV IRES-mediated translation, resulting in inhibition of HCV-IRES activity. Although overexpression of both NS3(pro) as well as the full length NS3 protein decreased the level of HCV IRES mediated translation, replication of HCV replicon RNA was enhanced significantly. These observations suggest that the NS3(pro) binding to HCV IRES reduces translation in favor of RNA replication. The competition between the host factor (La) and the viral protein (NS3) for binding to HCV IRES might regulate the molecular switch from translation to replication of HCV.
Figures

Similar articles
-
Inhibition of the interaction between NS3 protease and HCV IRES with a small peptide: a novel therapeutic strategy.Mol Ther. 2013 Jan;21(1):57-67. doi: 10.1038/mt.2012.151. Epub 2012 Aug 21. Mol Ther. 2013. PMID: 22910295 Free PMC article.
-
Human La protein interaction with GCAC near the initiator AUG enhances hepatitis C Virus RNA replication by promoting linkage between 5' and 3' untranslated regions.J Virol. 2013 Jun;87(12):6713-26. doi: 10.1128/JVI.00525-13. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552417 Free PMC article.
-
Protease Inhibitors Block Multiple Functions of the NS3/4A Protease-Helicase during the Hepatitis C Virus Life Cycle.J Virol. 2015 May;89(10):5362-70. doi: 10.1128/JVI.03188-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25740995 Free PMC article.
-
Functional and Physical Interaction between the Arf Activator GBF1 and Hepatitis C Virus NS3 Protein.J Virol. 2019 Mar 5;93(6):e01459-18. doi: 10.1128/JVI.01459-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567983 Free PMC article.
-
Hepatitis C Viral Replication Complex.Viruses. 2021 Mar 22;13(3):520. doi: 10.3390/v13030520. Viruses. 2021. PMID: 33809897 Free PMC article. Review.
Cited by
-
Human astroviruses: in silico analysis of the untranslated region and putative binding sites of cellular proteins.Mol Biol Rep. 2019 Feb;46(1):1413-1424. doi: 10.1007/s11033-018-4498-8. Epub 2018 Nov 17. Mol Biol Rep. 2019. PMID: 30448895 Free PMC article. Review.
-
Synergistic correlation between host angiogenin and dengue virus replication.RNA Biol. 2023 Jan;20(1):805-816. doi: 10.1080/15476286.2023.2264003. Epub 2023 Oct 5. RNA Biol. 2023. PMID: 37796112 Free PMC article.
-
A drug repurposing screen reveals dopamine signaling as a critical pathway underlying potential therapeutics for the rare disease DPAGT1-CDG.PLoS Genet. 2024 Oct 28;20(10):e1011458. doi: 10.1371/journal.pgen.1011458. eCollection 2024 Oct. PLoS Genet. 2024. PMID: 39466823 Free PMC article.
-
Identification and analysis of hepatitis C virus NS3 helicase inhibitors using nucleic acid binding assays.Nucleic Acids Res. 2012 Sep 1;40(17):8607-21. doi: 10.1093/nar/gks623. Epub 2012 Jun 27. Nucleic Acids Res. 2012. PMID: 22740655 Free PMC article.
-
Inhibition of the interaction between NS3 protease and HCV IRES with a small peptide: a novel therapeutic strategy.Mol Ther. 2013 Jan;21(1):57-67. doi: 10.1038/mt.2012.151. Epub 2012 Aug 21. Mol Ther. 2013. PMID: 22910295 Free PMC article.
References
-
- Barbato G. et al. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J. Mol. Biol. 289, 371–384 (1999). - PubMed
-
- Beran R. K. F., Serebrov V. & Pyle A. M. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 282, 34913–34920 (2007). - PubMed
-
- Ali N., Pruijn G. J. M., Kenan D. J., Keene J. D. & Siddiqqui A. Human La antigen is required for the Hepatitis C virus internal ribosome entry site mediated translation. J. Biol. Chem. 275, 27531–27540 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

