Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila
- PMID: 22354997
- PMCID: PMC3347239
- DOI: 10.1128/MCB.06503-11
Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila
Abstract
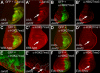
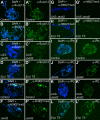
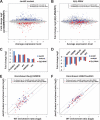
Jarid2 was recently identified as an important component of the mammalian Polycomb repressive complex 2 (PRC2), where it has a major effect on PRC2 recruitment in mouse embryonic stem cells. Although Jarid2 is conserved in Drosophila, it has not previously been implicated in Polycomb (Pc) regulation. Therefore, we purified Drosophila Jarid2 and its associated proteins and found that Jarid2 associates with all of the known canonical PRC2 components, demonstrating a conserved physical interaction with PRC2 in flies and mammals. Furthermore, in vivo studies with Jarid2 mutants in flies demonstrate that among several histone modifications tested, only methylation of histone 3 at K27 (H3K27), the mark implemented by PRC2, was affected. Genome-wide profiling of Jarid2, Su(z)12 (Suppressor of zeste 12), and H3K27me3 occupancy by chromatin immunoprecipitation with sequencing (ChIP-seq) indicates that Jarid2 and Su(z)12 have very similar distribution patterns on chromatin. However, Jarid2 and Su(z)12 occupancy levels at some genes are significantly different, with Jarid2 being present at relatively low levels at many Pc response elements (PREs) of certain Homeobox (Hox) genes, providing a rationale for why Jarid2 was never identified in Pc screens. Gene expression analyses show that Jarid2 and E(z) (Enhancer of zeste, a canonical PRC2 component) are not only required for transcriptional repression but might also function in active transcription. Identification of Jarid2 as a conserved PRC2 interactor in flies provides an opportunity to begin to probe some of its novel functions in Drosophila development.
Figures


Similar articles
-
Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation.Mol Cell. 2015 Mar 5;57(5):769-783. doi: 10.1016/j.molcel.2014.12.020. Epub 2015 Jan 22. Mol Cell. 2015. PMID: 25620564 Free PMC article.
-
Jarid2 and PRC2, partners in regulating gene expression.Genes Dev. 2010 Feb 15;24(4):368-80. doi: 10.1101/gad.1886410. Epub 2010 Feb 1. Genes Dev. 2010. PMID: 20123894 Free PMC article.
-
Elements of the polycomb repressor SU(Z)12 needed for histone H3-K27 methylation, the interface with E(Z), and in vivo function.Mol Cell Biol. 2013 Dec;33(24):4844-56. doi: 10.1128/MCB.00307-13. Epub 2013 Oct 7. Mol Cell Biol. 2013. PMID: 24100017 Free PMC article.
-
Inner workings and regulatory inputs that control Polycomb repressive complex 2.Chromosoma. 2012 Jun;121(3):221-34. doi: 10.1007/s00412-012-0361-1. Epub 2012 Feb 19. Chromosoma. 2012. PMID: 22349693 Free PMC article. Review.
-
Recruiting polycomb to chromatin.Int J Biochem Cell Biol. 2015 Oct;67:177-87. doi: 10.1016/j.biocel.2015.05.006. Epub 2015 May 14. Int J Biochem Cell Biol. 2015. PMID: 25982201 Free PMC article. Review.
Cited by
-
Polycomb Repressive Complex 2 in Eukaryotes-An Evolutionary Perspective.Epigenomes. 2022 Jan 17;6(1):3. doi: 10.3390/epigenomes6010003. Epigenomes. 2022. PMID: 35076495 Free PMC article. Review.
-
A bioinformatics screen reveals hox and chromatin remodeling factors at the Drosophila histone locus.BMC Genom Data. 2023 Sep 21;24(1):54. doi: 10.1186/s12863-023-01147-0. BMC Genom Data. 2023. PMID: 37735352 Free PMC article.
-
Mapping the Phosphorylation Pattern of Drosophila melanogaster RNA Polymerase II Carboxyl-Terminal Domain Using Ultraviolet Photodissociation Mass Spectrometry.ACS Chem Biol. 2017 Jan 20;12(1):153-162. doi: 10.1021/acschembio.6b00729. Epub 2016 Dec 1. ACS Chem Biol. 2017. PMID: 28103682 Free PMC article.
-
Polycomb silencing of the Drosophila 4E-BP gene regulates imaginal disc cell growth.Dev Biol. 2013 Aug 1;380(1):111-24. doi: 10.1016/j.ydbio.2013.03.011. Epub 2013 Mar 20. Dev Biol. 2013. PMID: 23523430 Free PMC article.
-
PRC2.1- and PRC2.2-specific accessory proteins drive recruitment of different forms of canonical PRC1.Mol Cell. 2023 May 4;83(9):1393-1411.e7. doi: 10.1016/j.molcel.2023.03.018. Epub 2023 Apr 7. Mol Cell. 2023. PMID: 37030288 Free PMC article.
References
-
- Beisel C, Paro R. 2011. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12:123–135 - PubMed
-
- Boyer LA, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 - PubMed
-
- Cao R, Zhang Y. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15:57–67 - PubMed
-
- Carrington EA, Jones RS. 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development 122:4073–4083 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
