IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype
- PMID: 22343889
- PMCID: PMC3351699
- DOI: 10.1038/nature10866
IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype
Abstract
Both genome-wide genetic and epigenetic alterations are fundamentally important for the development of cancers, but the interdependence of these aberrations is poorly understood. Glioblastomas and other cancers with the CpG island methylator phenotype (CIMP) constitute a subset of tumours with extensive epigenomic aberrations and a distinct biology. Glioma CIMP (G-CIMP) is a powerful determinant of tumour pathogenicity, but the molecular basis of G-CIMP remains unresolved. Here we show that mutation of a single gene, isocitrate dehydrogenase 1 (IDH1), establishes G-CIMP by remodelling the methylome. This remodelling results in reorganization of the methylome and transcriptome. Examination of the epigenome of a large set of intermediate-grade gliomas demonstrates a distinct G-CIMP phenotype that is highly dependent on the presence of IDH mutation. Introduction of mutant IDH1 into primary human astrocytes alters specific histone marks, induces extensive DNA hypermethylation, and reshapes the methylome in a fashion that mirrors the changes observed in G-CIMP-positive lower-grade gliomas. Furthermore, the epigenomic alterations resulting from mutant IDH1 activate key gene expression programs, characterize G-CIMP-positive proneural glioblastomas but not other glioblastomas, and are predictive of improved survival. Our findings demonstrate that IDH mutation is the molecular basis of CIMP in gliomas, provide a framework for understanding oncogenesis in these gliomas, and highlight the interplay between genomic and epigenomic changes in human cancers.
Figures

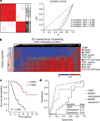
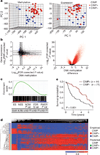
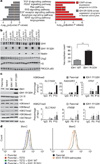
Comment in
-
Metabolism: unmasking an oncometabolite.Nat Rev Cancer. 2012 Mar 1;12(4):229. doi: 10.1038/nrc3248. Nat Rev Cancer. 2012. PMID: 22378191 No abstract available.
Similar articles
-
CRISPR Editing of Mutant IDH1 R132H Induces a CpG Methylation-Low State in Patient-Derived Glioma Models of G-CIMP.Mol Cancer Res. 2019 Oct;17(10):2042-2050. doi: 10.1158/1541-7786.MCR-19-0309. Epub 2019 Jul 10. Mol Cancer Res. 2019. PMID: 31292202 Free PMC article.
-
Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma.Cancer Cell. 2010 May 18;17(5):510-22. doi: 10.1016/j.ccr.2010.03.017. Epub 2010 Apr 15. Cancer Cell. 2010. PMID: 20399149 Free PMC article.
-
A Distinct DNA Methylation Shift in a Subset of Glioma CpG Island Methylator Phenotypes during Tumor Recurrence.Cell Rep. 2018 Apr 10;23(2):637-651. doi: 10.1016/j.celrep.2018.03.107. Cell Rep. 2018. PMID: 29642018 Free PMC article.
-
Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications.Neuro Oncol. 2018 Apr 9;20(5):608-620. doi: 10.1093/neuonc/nox183. Neuro Oncol. 2018. PMID: 29036500 Free PMC article. Review.
-
Overview of DNA methylation in adult diffuse gliomas.Brain Tumor Pathol. 2019 Apr;36(2):84-91. doi: 10.1007/s10014-019-00339-w. Epub 2019 Apr 1. Brain Tumor Pathol. 2019. PMID: 30937703 Review.
Cited by
-
Methylation Profiling in Diffuse Gliomas: Diagnostic Value and Considerations.Cancers (Basel). 2022 Nov 18;14(22):5679. doi: 10.3390/cancers14225679. Cancers (Basel). 2022. PMID: 36428772 Free PMC article. Review.
-
What do we know about IDH1/2 mutations so far, and how do we use it?Acta Neuropathol. 2013 May;125(5):621-36. doi: 10.1007/s00401-013-1106-9. Epub 2013 Mar 20. Acta Neuropathol. 2013. PMID: 23512379 Free PMC article. Review.
-
Chemical approaches to study metabolic networks.Pflugers Arch. 2013 Mar;465(3):427-40. doi: 10.1007/s00424-012-1201-0. Epub 2013 Jan 8. Pflugers Arch. 2013. PMID: 23296751 Free PMC article. Review.
-
Molecularly targeted therapies in non-small-cell lung cancer annual update 2014.J Thorac Oncol. 2015 Jan;10(1 Suppl 1):S1-63. doi: 10.1097/JTO.0000000000000405. J Thorac Oncol. 2015. PMID: 25535693 Free PMC article.
-
MethylMeter(®): bisulfite-free quantitative and sensitive DNA methylation profiling and mutation detection in FFPE samples.Epigenomics. 2016 Jun;8(6):747-65. doi: 10.2217/epi-2016-0004. Epub 2016 Jun 23. Epigenomics. 2016. PMID: 27337298 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

