JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis
- PMID: 22327740
- PMCID: PMC3315231
- DOI: 10.1105/tpc.111.093005
JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis
Abstract
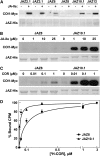
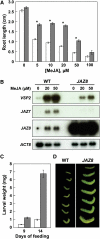
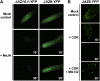
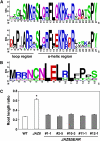
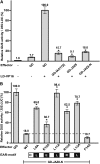
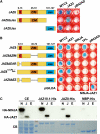
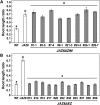
The lipid-derived hormone jasmonoyl-L-Ile (JA-Ile) initiates large-scale changes in gene expression by stabilizing the interaction of JASMONATE ZIM domain (JAZ) repressors with the F-box protein CORONATINE INSENSITIVE1 (COI1), which results in JAZ degradation by the ubiquitin-proteasome pathway. Recent structural studies show that the JAZ1 degradation signal (degron) includes a short conserved LPIAR motif that seals JA-Ile in its binding pocket at the COI1-JAZ interface. Here, we show that Arabidopsis thaliana JAZ8 lacks this motif and thus is unable to associate strongly with COI1 in the presence of JA-Ile. As a consequence, JAZ8 is stabilized against jasmonate (JA)-mediated degradation and, when ectopically expressed in Arabidopsis, represses JA-regulated growth and defense responses. These findings indicate that sequence variation in a hypervariable region of the degron affects JAZ stability and JA-regulated physiological responses. We also show that JAZ8-mediated repression depends on an LxLxL-type EAR (for ERF-associated amphiphilic repression) motif at the JAZ8 N terminus that binds the corepressor TOPLESS and represses transcriptional activation. JAZ8-mediated repression does not require the ZIM domain, which, in other JAZ proteins, recruits TOPLESS through the EAR motif-containing adaptor protein NINJA. These findings show that EAR repression domains in a subgroup of JAZ proteins repress gene expression through direct recruitment of corepressors to cognate transcription factors.
Figures


Similar articles
-
A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein.Plant J. 2008 Sep;55(6):979-88. doi: 10.1111/j.1365-313X.2008.03566.x. Epub 2008 Jun 10. Plant J. 2008. PMID: 18547396 Free PMC article.
-
A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.Plant Cell. 2009 Jan;21(1):131-45. doi: 10.1105/tpc.108.064097. Epub 2009 Jan 16. Plant Cell. 2009. PMID: 19151223 Free PMC article.
-
Arabidopsis ECAP Is a New Adaptor Protein that Connects JAZ Repressors with the TPR2 Co-repressor to Suppress Jasmonate-Responsive Anthocyanin Accumulation.Mol Plant. 2020 Feb 3;13(2):246-265. doi: 10.1016/j.molp.2019.10.014. Epub 2019 Nov 6. Mol Plant. 2020. PMID: 31706031
-
The JAZ proteins: a crucial interface in the jasmonate signaling cascade.Plant Cell. 2011 Sep;23(9):3089-100. doi: 10.1105/tpc.111.089300. Epub 2011 Sep 30. Plant Cell. 2011. PMID: 21963667 Free PMC article. Review.
-
Arabidopsis jasmonate signaling pathway.Sci Signal. 2010 Feb 16;3(109):cm4. doi: 10.1126/scisignal.3109cm4. Sci Signal. 2010. PMID: 20159850 Review.
Cited by
-
Jasmonates and Histone deacetylase 6 activate Arabidopsis genome-wide histone acetylation and methylation during the early acute stress response.BMC Biol. 2022 Apr 11;20(1):83. doi: 10.1186/s12915-022-01273-8. BMC Biol. 2022. PMID: 35399062 Free PMC article.
-
Jasmonate Zim-Domain Protein 9 Interacts With Slender Rice 1 to Mediate the Antagonistic Interaction Between Jasmonic and Gibberellic Acid Signals in Rice.Front Plant Sci. 2018 Dec 18;9:1866. doi: 10.3389/fpls.2018.01866. eCollection 2018. Front Plant Sci. 2018. PMID: 30619427 Free PMC article.
-
CUL3BPM E3 ubiquitin ligases regulate MYC2, MYC3, and MYC4 stability and JA responses.Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):6205-6215. doi: 10.1073/pnas.1912199117. Epub 2020 Mar 2. Proc Natl Acad Sci U S A. 2020. PMID: 32123086 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of JAZ Family Involved in Hormone and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2021 Sep 10;22(18):9786. doi: 10.3390/ijms22189786. Int J Mol Sci. 2021. PMID: 34575953 Free PMC article.
-
Hormone crosstalk in wound stress response: wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana.J Exp Bot. 2016 Mar;67(7):2107-20. doi: 10.1093/jxb/erv521. Epub 2015 Dec 15. J Exp Bot. 2016. PMID: 26672615 Free PMC article.
References
-
- Bai Y., Meng Y., Huang D., Qi Y., Chen M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136 - PubMed
-
- Ballaré C.L. (2011). Jasmonate-induced defenses: A tale of intelligence, collaborators and rascals. Trends Plant Sci. 16: 249–257 - PubMed
-
- Blechert S., Bockelmann C., Fusslein M., Von Schrader T., Stelmach B., Niesel U., Weiler E.W. (1999). Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207: 470–479
-
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

