Overexpression of the DNA sensor proteins, absent in melanoma 2 and interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation
- PMID: 22320325
- PMCID: PMC7659186
- DOI: 10.1111/j.1349-7006.2012.02211.x
Overexpression of the DNA sensor proteins, absent in melanoma 2 and interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation
Abstract
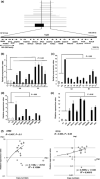
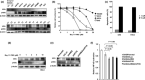
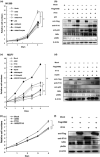
The development of oral squamous cell carcinoma (OSCC) is a multistep process that requires the accumulation of genetic alterations. To identify genes responsible for OSCC development, we performed high-density single nucleotide polymorphism array analysis and genome-wide gene expression profiling on OSCC tumors. These analyses indicated that the absent in melanoma 2 (AIM2) gene and the interferon-inducible gene 16 (IFI16) mapped to the hematopoietic interferon-inducible nuclear proteins. The 200-amino-acid repeat gene cluster in the amplified region of chromosome 1q23 is overexpressed in OSCC. Both AIM2 and IFI16 are cytoplasmic double-stranded DNA sensors for innate immunity and act as tumor suppressors in several human cancers. Knockdown of AIM2 or IFI16 in OSCC cells results in the suppression of cell growth and apoptosis, accompanied by the downregulation of nuclear factor kappa-light-chain-enhancer of activated B cells activation. Because all OSCC cell lines have reduced p53 activity, wild-type p53 was introduced in p53-deficient OSCC cells. The expression of wild-type p53 suppressed cell growth and induced apoptosis via suppression of nuclear factor kappa-light-chain-enhancer of activated B cells activity. Finally, the co-expression of AIM2 and IFI16 significantly enhanced cell growth in p53-deficient cells; in contrast, the expression of AIM2 and/or IFI16 in cells bearing wild-type p53 suppressed cell growth. Moreover, AIM2 and IFI16 synergistically enhanced nuclear factor kappa-light-chain-enhancer of activated B cells signaling in p53-deficient cells. Thus, expression of AIM2 and IFI16 may have oncogenic activities in the OSCC cells that have inactivated the p53 system.
© 2012 Japanese Cancer Association.
Figures


Similar articles
-
Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts.Mol Cancer Res. 2011 May;9(5):589-602. doi: 10.1158/1541-7786.MCR-10-0565. Epub 2011 Apr 6. Mol Cancer Res. 2011. PMID: 21471287 Free PMC article.
-
Overexpression of absent in melanoma 2 in oral squamous cell carcinoma contributes to tumor progression.Biochem Biophys Res Commun. 2019 Jan 29;509(1):82-88. doi: 10.1016/j.bbrc.2018.12.066. Epub 2018 Dec 23. Biochem Biophys Res Commun. 2019. PMID: 30587341
-
AIM2 promotes irradiation resistance, migration ability and PD-L1 expression through STAT1/NF-κB activation in oral squamous cell carcinoma.J Transl Med. 2024 Jan 3;22(1):13. doi: 10.1186/s12967-023-04825-w. J Transl Med. 2024. PMID: 38166970 Free PMC article.
-
Effects of AIM2 and IFI16 on Infectious Diseases and Inflammation.Viral Immunol. 2023 Sep;36(7):438-448. doi: 10.1089/vim.2023.0044. Epub 2023 Aug 16. Viral Immunol. 2023. PMID: 37585649 Review.
-
Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization.Mol Immunol. 2012 Jan;49(4):567-71. doi: 10.1016/j.molimm.2011.11.004. Epub 2011 Dec 2. Mol Immunol. 2012. PMID: 22137500 Free PMC article. Review.
Cited by
-
AIM2 Inflammasome's First Decade of Discovery: Focus on Oral Diseases.Front Immunol. 2020 Aug 13;11:1487. doi: 10.3389/fimmu.2020.01487. eCollection 2020. Front Immunol. 2020. PMID: 32903550 Free PMC article. Review.
-
PYHIN Proteins and HPV: Role in the Pathogenesis of Head and Neck Squamous Cell Carcinoma.Microorganisms. 2019 Dec 20;8(1):14. doi: 10.3390/microorganisms8010014. Microorganisms. 2019. PMID: 31861809 Free PMC article. Review.
-
Increased expression of IFI16 predicts adverse prognosis in multiple myeloma.Pharmacogenomics J. 2021 Aug;21(4):520-532. doi: 10.1038/s41397-021-00230-y. Epub 2021 Mar 12. Pharmacogenomics J. 2021. PMID: 33712724
-
Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review.Cancers (Basel). 2023 Nov 27;15(23):5600. doi: 10.3390/cancers15235600. Cancers (Basel). 2023. PMID: 38067304 Free PMC article. Review.
-
Absent in melanoma 2 suppresses gastric cancer cell proliferation and migration via inactivation of AKT signaling pathway.Sci Rep. 2021 Apr 15;11(1):8235. doi: 10.1038/s41598-021-87744-4. Sci Rep. 2021. PMID: 33859277 Free PMC article.
References
-
- Shah JP, Singh B. Keynote comment: why the lack of progress for oral cancer? Lancet Oncol 2006; 7: 356–7. - PubMed
-
- Gibson MK, Forastiere AA. Reassessment of the role of induction chemotherapy for head and neck cancer. Lancet Oncol 2006; 7: 565–74. - PubMed
-
- Perez‐Sayans M, Somoza‐Martin JM, Barros‐Angueira F, Reboiras‐Lopez MD, Gandara Rey JM, Garcia‐Garcia A. Genetic and molecular alterations associated with oral squamous cell cancer (Review). Oncol Rep 2009; 22: 1277–82. - PubMed
-
- Rogers SJ, Harrington KJ, Rhys‐Evans P, P OC, Eccles SA. Biological significance of c‐erbB family oncogenes in head and neck cancer. Cancer Metastasis Rev 2005; 24: 47–69. - PubMed
-
- Rousseau A, Lim MS, Lin Z, Jordan RC. Frequent cyclin D1 gene amplification and protein overexpression in oral epithelial dysplasias. Oral Oncol 2001; 37: 268–75. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

