Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments
- PMID: 22318319
- PMCID: PMC3280463
- DOI: 10.1128/mBio.00301-11
Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments
Abstract
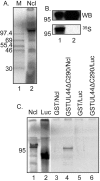
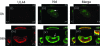
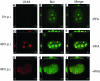
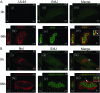
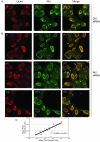
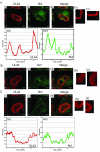
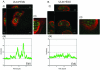
Drastic reorganization of the nucleus is a hallmark of herpesvirus replication. This reorganization includes the formation of viral replication compartments, the subnuclear structures in which the viral DNA genome is replicated. The architecture of replication compartments is poorly understood. However, recent work with human cytomegalovirus (HCMV) showed that the viral DNA polymerase subunit UL44 concentrates and viral DNA synthesis occurs at the periphery of these compartments. Any cellular factors involved in replication compartment architecture are largely unknown. Previously, we found that nucleolin, a major protein component of nucleoli, associates with HCMV UL44 in infected cells and is required for efficient viral DNA synthesis. Here, we show that nucleolin binds to purified UL44. Confocal immunofluorescence analysis demonstrated colocalization of nucleolin with UL44 at the periphery of replication compartments. Pharmacological inhibition of viral DNA synthesis prevented the formation of replication compartments but did not abrogate association of UL44 and nucleolin. Thus, association of UL44 and nucleolin is unlikely to be a nonspecific effect related to development of replication compartments. No detectable colocalization of 5-ethynyl-2'-deoxyuridine (EdU)-labeled viral DNA with nucleolin was observed, suggesting that nucleolin is not directly involved in viral DNA synthesis. Small interfering RNA (siRNA)-mediated knockdown of nucleolin caused improper localization of UL44 and a defect in EdU incorporation into viral DNA. We propose a model in which nucleolin anchors UL44 at the periphery of replication compartments to maintain their architecture and promote viral DNA synthesis.
Importance: Human cytomegalovirus (HCMV) is an important human pathogen. HCMV infection causes considerable rearrangement of the structure of the nucleus, largely due to the formation of viral replication compartments within the nucleus. Within these compartments, the virus replicates its DNA genome. We previously demonstrated that nucleolin is required for efficient viral DNA synthesis and now find that the nucleolar protein nucleolin interacts with a subunit of the viral DNA polymerase, UL44, specifically at the periphery of replication compartments. Moreover, we find that nucleolin is required to properly localize UL44 at this region. Nucleolin is, therefore, involved in the organization of proteins within replication compartments. This, to our knowledge, is the first report identifying a cellular protein required for maintaining replication compartment architecture.
Figures

Similar articles
-
Dynamic and nucleolin-dependent localization of human cytomegalovirus UL84 to the periphery of viral replication compartments and nucleoli.J Virol. 2014 Oct;88(20):11738-47. doi: 10.1128/JVI.01889-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078694 Free PMC article.
-
Human cytomegalovirus UL44 concentrates at the periphery of replication compartments, the site of viral DNA synthesis.J Virol. 2012 Feb;86(4):2089-95. doi: 10.1128/JVI.06720-11. Epub 2011 Dec 7. J Virol. 2012. PMID: 22156516 Free PMC article.
-
Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication.J Virol. 2010 Feb;84(4):1771-84. doi: 10.1128/JVI.01510-09. Epub 2009 Dec 9. J Virol. 2010. PMID: 20007282 Free PMC article.
-
Viral and cellular subnuclear structures in human cytomegalovirus-infected cells.J Gen Virol. 2015 Feb;96(Pt 2):239-252. doi: 10.1099/vir.0.071084-0. Epub 2014 Oct 30. J Gen Virol. 2015. PMID: 25359764 Review.
-
Nuts and bolts of human cytomegalovirus lytic DNA replication.Curr Top Microbiol Immunol. 2008;325:153-66. doi: 10.1007/978-3-540-77349-8_9. Curr Top Microbiol Immunol. 2008. PMID: 18637505 Review.
Cited by
-
Dissecting the Role of DDX21 in Regulating Human Cytomegalovirus Replication.J Virol. 2019 Nov 26;93(24):e01222-19. doi: 10.1128/JVI.01222-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554690 Free PMC article.
-
Seneca Valley Virus 3Cpro Mediates Cleavage and Redistribution of Nucleolin To Facilitate Viral Replication.Microbiol Spectr. 2022 Apr 27;10(2):e0030422. doi: 10.1128/spectrum.00304-22. Epub 2022 Mar 31. Microbiol Spectr. 2022. PMID: 35357201 Free PMC article.
-
Dynamic and nucleolin-dependent localization of human cytomegalovirus UL84 to the periphery of viral replication compartments and nucleoli.J Virol. 2014 Oct;88(20):11738-47. doi: 10.1128/JVI.01889-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078694 Free PMC article.
-
Variation in the nuclear effects of infection by different human rhinovirus serotypes.Front Microbiol. 2015 Aug 24;6:875. doi: 10.3389/fmicb.2015.00875. eCollection 2015. Front Microbiol. 2015. PMID: 26379650 Free PMC article.
-
Morphological, Biochemical, and Functional Study of Viral Replication Compartments Isolated from Adenovirus-Infected Cells.J Virol. 2016 Jan 13;90(7):3411-27. doi: 10.1128/JVI.00033-16. J Virol. 2016. PMID: 26764008 Free PMC article.
References
-
- Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
