NGS QC Toolkit: a toolkit for quality control of next generation sequencing data
- PMID: 22312429
- PMCID: PMC3270013
- DOI: 10.1371/journal.pone.0030619
NGS QC Toolkit: a toolkit for quality control of next generation sequencing data
Abstract
Next generation sequencing (NGS) technologies provide a high-throughput means to generate large amount of sequence data. However, quality control (QC) of sequence data generated from these technologies is extremely important for meaningful downstream analysis. Further, highly efficient and fast processing tools are required to handle the large volume of datasets. Here, we have developed an application, NGS QC Toolkit, for quality check and filtering of high-quality data. This toolkit is a standalone and open source application freely available at http://www.nipgr.res.in/ngsqctoolkit.html. All the tools in the application have been implemented in Perl programming language. The toolkit is comprised of user-friendly tools for QC of sequencing data generated using Roche 454 and Illumina platforms, and additional tools to aid QC (sequence format converter and trimming tools) and analysis (statistics tools). A variety of options have been provided to facilitate the QC at user-defined parameters. The toolkit is expected to be very useful for the QC of NGS data to facilitate better downstream analysis.
Conflict of interest statement
Figures
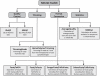
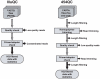

Similar articles
-
QC-Chain: fast and holistic quality control method for next-generation sequencing data.PLoS One. 2013;8(4):e60234. doi: 10.1371/journal.pone.0060234. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565205 Free PMC article.
-
Software for pre-processing Illumina next-generation sequencing short read sequences.Source Code Biol Med. 2014 May 3;9:8. doi: 10.1186/1751-0473-9-8. eCollection 2014. Source Code Biol Med. 2014. PMID: 24955109 Free PMC article.
-
ClinQC: a tool for quality control and cleaning of Sanger and NGS data in clinical research.BMC Bioinformatics. 2016 Feb 2;17:56. doi: 10.1186/s12859-016-0915-y. BMC Bioinformatics. 2016. PMID: 26830926 Free PMC article.
-
Prevention, diagnosis and treatment of high-throughput sequencing data pathologies.Mol Ecol. 2014 Apr;23(7):1679-700. doi: 10.1111/mec.12680. Epub 2014 Mar 13. Mol Ecol. 2014. PMID: 24471475 Review.
-
Standardization and quality management in next-generation sequencing.Appl Transl Genom. 2016 Jul 1;10:2-9. doi: 10.1016/j.atg.2016.06.001. eCollection 2016 Sep. Appl Transl Genom. 2016. PMID: 27668169 Free PMC article. Review.
Cited by
-
Temporal Changes of Virus-Like Particle Abundance and Metagenomic Comparison of Viral Communities in Cropland and Prairie Soils.mSphere. 2021 Jun 30;6(3):e0116020. doi: 10.1128/mSphere.01160-20. Epub 2021 Jun 2. mSphere. 2021. PMID: 34077260 Free PMC article.
-
Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae).Plant Divers. 2020 Jul 25;43(1):15-26. doi: 10.1016/j.pld.2020.07.004. eCollection 2021 Feb. Plant Divers. 2020. PMID: 33778221 Free PMC article.
-
Effects of Nitrate Addition on Rumen Fermentation, Bacterial Biodiversity and Abundance.Asian-Australas J Anim Sci. 2015 Oct;28(10):1433-41. doi: 10.5713/ajas.15.0091. Asian-Australas J Anim Sci. 2015. PMID: 26194220 Free PMC article.
-
Genomic Analysis Reveals Candidate Genes Underlying Sex-Linked Eyelid Coloboma, Feather Color Traits, and Climatic Adaptation in Huoyan Geese.Animals (Basel). 2023 Nov 22;13(23):3608. doi: 10.3390/ani13233608. Animals (Basel). 2023. PMID: 38066959 Free PMC article.
-
Data Mining of Lung Microbiota in Cystic Fibrosis Patients.PLoS One. 2016 Oct 14;11(10):e0164510. doi: 10.1371/journal.pone.0164510. eCollection 2016. PLoS One. 2016. PMID: 27741283 Free PMC article.
References
-
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

