Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells
- PMID: 22309939
- PMCID: PMC3496140
- DOI: 10.1186/bcr3107
Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells
Abstract
Introduction: Triple-negative breast cancer (TNBC) high rate of relapse is thought to be due to the presence of tumor-initiating cells (TICs), molecularly defined as being CD44high/CD24-/low. TICs are resilient to chemotherapy and radiation. However, no currently accepted molecular target exists against TNBC and, moreover, TICs. Therefore, we sought the identification of kinase targets that inhibit TNBC growth and eliminate TICs.
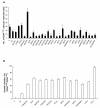
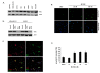
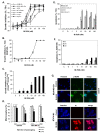
Methods: A genome-wide human kinase small interfering RNA (siRNA) library (691 kinases) was screened against the TNBC cell line SUM149 for growth inhibition. Selected siRNAs were then tested on four different breast cancer cell lines to confirm the spectrum of activity. Their effect on the CD44high subpopulation and sorted CD44high/CD24-/low cells of SUM149 also was studied. Further studies were focused on polo-like kinase 1 (PLK1), including its expression in breast cancer cell lines, effect on the CD44high/CD24-/low TIC subpopulation, growth inhibition, mammosphere formation, and apoptosis, as well as the activity of the PLK1 inhibitor, BI 2536.
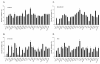
Results: Of the 85 kinases identified in the screen, 28 of them were further silenced by siRNAs on MDA-MB-231 (TNBC), BT474-M1 (ER+/HER2+, a metastatic variant), and HR5 (ER+/HER2+, a trastuzumab-resistant model) cells and showed a broad spectrum of growth inhibition. Importantly, 12 of 28 kinases also reduced the CD44high subpopulation compared with control in SUM149. Further tests of these 12 kinases directly on a sorted CD44high/CD24-/low TIC subpopulation of SUM149 cells confirmed their effect. Blocking PLK1 had the greatest growth inhibition on breast cancer cells and TICs by about 80% to 90% after 72 hours. PLK1 was universally expressed in breast cancer cell lines, representing all of the breast cancer subtypes, and was positively correlated to CD44. The PLK1 inhibitor BI 2536 showed similar effects on growth, mammosphere formation, and apoptosis as did PLK1 siRNAs. Finally, whereas paclitaxel, doxorubicin, and 5-fluorouracil enriched the CD44high/CD24-/low population compared with control in SUM149, subsequent treatment with BI 2536 killed the emergent population, suggesting that it could potentially be used to prevent relapse.
Conclusion: Inhibiting PLK1 with siRNA or BI 2536 blocked growth of TNBCs including the CD44high/CD24-/low TIC subpopulation and mammosphere formation. Thus, PLK1 could be a potential therapeutic target for the treatment of TNBC as well as other subtypes of breast cancer.
Figures
Similar articles
-
Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer.Cancer Res. 2013 Jan 15;73(2):813-23. doi: 10.1158/0008-5472.CAN-12-2633. Epub 2012 Nov 9. Cancer Res. 2013. PMID: 23144294
-
Therapeutic potential of PLK1 inhibition in triple-negative breast cancer.Lab Invest. 2019 Sep;99(9):1275-1286. doi: 10.1038/s41374-019-0247-4. Epub 2019 Apr 17. Lab Invest. 2019. PMID: 30996295
-
Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer.Cancer Res. 2015 Jan 15;75(2):405-14. doi: 10.1158/0008-5472.CAN-14-2475. Epub 2014 Dec 5. Cancer Res. 2015. PMID: 25480943 Free PMC article.
-
Plk1-targeted small molecule inhibitors: molecular basis for their potency and specificity.Mol Cells. 2011 Sep;32(3):209-20. doi: 10.1007/s10059-011-0126-3. Epub 2011 Jul 29. Mol Cells. 2011. PMID: 21809214 Free PMC article. Review.
-
Polo-like Kinase 1 as an emerging drug target: structure, function and therapeutic implications.J Drug Target. 2021 Feb;29(2):168-184. doi: 10.1080/1061186X.2020.1818760. Epub 2020 Sep 14. J Drug Target. 2021. PMID: 32886539 Review.
Cited by
-
Dual TTK/PLK1 inhibition has potent anticancer activity in TNBC as monotherapy and in combination.Front Oncol. 2024 Aug 9;14:1447807. doi: 10.3389/fonc.2024.1447807. eCollection 2024. Front Oncol. 2024. PMID: 39184047 Free PMC article.
-
Therapeutic targeting of polo-like kinase 1 using RNA-interfering nanoparticles (iNOPs) for the treatment of non-small cell lung cancer.Oncotarget. 2015 May 20;6(14):12020-34. doi: 10.18632/oncotarget.2664. Oncotarget. 2015. PMID: 25557168 Free PMC article.
-
Triple-negative Breast Cancer: Identification of circRNAs With Efficacy in Preclinical In Vivo Models.Cancer Genomics Proteomics. 2023 Mar-Apr;20(2):117-131. doi: 10.21873/cgp.20368. Cancer Genomics Proteomics. 2023. PMID: 36870692 Free PMC article. Review.
-
Identification of key genes controlling breast cancer stem cell characteristics via stemness indices analysis.J Transl Med. 2020 Feb 12;18(1):74. doi: 10.1186/s12967-020-02260-9. J Transl Med. 2020. PMID: 32050983 Free PMC article.
-
PLK1, A Potential Target for Cancer Therapy.Transl Oncol. 2017 Feb;10(1):22-32. doi: 10.1016/j.tranon.2016.10.003. Epub 2016 Nov 24. Transl Oncol. 2017. PMID: 27888710 Free PMC article. Review.
References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenchikov A, Williams C, Zhu SX, Lønning PE, Børreson-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

