Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis
- PMID: 22296834
- PMCID: PMC3381520
- DOI: 10.1126/scisignal.2002446
Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis
Abstract
Class I myosins participate in various interactions between the cell membrane and the cytoskeleton. Several class I myosins preferentially bind to acidic phospholipids, such as phosphatidylserine and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], through a tail homology 1 (TH1) domain. Here, we show that the second messenger lipid phosphatidylinositol 3,4,5-trisphosphate (PIP3) binds to the TH1 domain of a subset of Dictyostelium class I myosins (ID, IE, and IF) and recruits them to the plasma membrane. The PIP3-regulated membrane recruitment of myosin I promoted chemotaxis and induced chemoattractant-stimulated actin polymerization. Similarly, PIP3 recruited human myosin IF to the plasma membrane upon chemotactic stimulation in a neutrophil cell line. These data suggest a mechanism through which the PIP3 signal is transmitted through myosin I to the actin cytoskeleton.
Figures
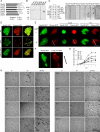
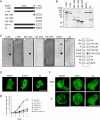
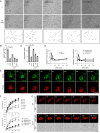
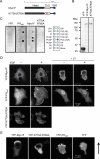
Similar articles
-
Molecular basis of dynamic relocalization of Dictyostelium myosin IB.J Biol Chem. 2012 Apr 27;287(18):14923-36. doi: 10.1074/jbc.M111.318667. Epub 2012 Feb 24. J Biol Chem. 2012. PMID: 22367211 Free PMC article.
-
Myosin I: A new pip(3) effector in chemotaxis and phagocytosis.Commun Integr Biol. 2012 May 1;5(3):294-6. doi: 10.4161/cib.19892. Commun Integr Biol. 2012. PMID: 22896797 Free PMC article.
-
Generation of cells that ignore the effects of PIP3 on cytoskeleton.Cell Cycle. 2011 Sep 1;10(17):2817-8. doi: 10.4161/cc.10.17.16744. Epub 2011 Sep 1. Cell Cycle. 2011. PMID: 21869609 Free PMC article. No abstract available.
-
Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling.Cell Signal. 2015 Sep;27(9):1789-98. doi: 10.1016/j.cellsig.2015.05.013. Epub 2015 May 27. Cell Signal. 2015. PMID: 26022180 Review.
-
Regulation of Dictyostelium myosin I and II.Biochim Biophys Acta. 2001 Mar 15;1525(3):245-61. doi: 10.1016/s0304-4165(01)00110-6. Biochim Biophys Acta. 2001. PMID: 11257438 Review.
Cited by
-
IQGAP-related protein IqgC suppresses Ras signaling during large-scale endocytosis.Proc Natl Acad Sci U S A. 2019 Jan 22;116(4):1289-1298. doi: 10.1073/pnas.1810268116. Epub 2019 Jan 8. Proc Natl Acad Sci U S A. 2019. PMID: 30622175 Free PMC article.
-
Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes.Cell Mol Life Sci. 2014 Oct;71(19):3711-47. doi: 10.1007/s00018-014-1638-8. Epub 2014 May 21. Cell Mol Life Sci. 2014. PMID: 24846395 Free PMC article. Review.
-
Class I myosins have overlapping and specialized functions in left-right asymmetric development in Drosophila.Genetics. 2015 Apr;199(4):1183-99. doi: 10.1534/genetics.115.174698. Epub 2015 Feb 6. Genetics. 2015. PMID: 25659376 Free PMC article.
-
PakB binds to the SH3 domain of Dictyostelium Abp1 and regulates its effects on cell polarity and early development.Mol Biol Cell. 2013 Jul;24(14):2216-27. doi: 10.1091/mbc.E12-12-0883. Epub 2013 May 22. Mol Biol Cell. 2013. PMID: 23699396 Free PMC article.
-
A new class of cancer-associated PTEN mutations defined by membrane translocation defects.Oncogene. 2015 Jul;34(28):3737-43. doi: 10.1038/onc.2014.293. Epub 2014 Sep 29. Oncogene. 2015. PMID: 25263454 Free PMC article.
References
-
- Van Haastert PJ. Chemotaxis: insights from the extending pseudopod. J Cell Sci. 2010;123:3031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

