JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data
- PMID: 22285562
- PMCID: PMC3315723
- DOI: 10.1093/bioinformatics/bts053
JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data
Abstract
Motivation: Identification of somatic single nucleotide variants (SNVs) in tumour genomes is a necessary step in defining the mutational landscapes of cancers. Experimental designs for genome-wide ascertainment of somatic mutations now routinely include next-generation sequencing (NGS) of tumour DNA and matched constitutional DNA from the same individual. This allows investigators to control for germline polymorphisms and distinguish somatic mutations that are unique to the tumour, thus reducing the burden of labour-intensive and expensive downstream experiments needed to verify initial predictions. In order to make full use of such paired datasets, computational tools for simultaneous analysis of tumour-normal paired sequence data are required, but are currently under-developed and under-represented in the bioinformatics literature.
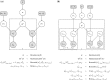
Results: In this contribution, we introduce two novel probabilistic graphical models called JointSNVMix1 and JointSNVMix2 for jointly analysing paired tumour-normal digital allelic count data from NGS experiments. In contrast to independent analysis of the tumour and normal data, our method allows statistical strength to be borrowed across the samples and therefore amplifies the statistical power to identify and distinguish both germline and somatic events in a unified probabilistic framework.
Availability: The JointSNVMix models and four other models discussed in the article are part of the JointSNVMix software package available for download at http://compbio.bccrc.ca
Contact: sshah@bccrc.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures


Similar articles
-
multiSNV: a probabilistic approach for improving detection of somatic point mutations from multiple related tumour samples.Nucleic Acids Res. 2015 May 19;43(9):e61. doi: 10.1093/nar/gkv135. Epub 2015 Feb 26. Nucleic Acids Res. 2015. PMID: 25722372 Free PMC article.
-
Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data.Bioinformatics. 2012 Jan 15;28(2):167-75. doi: 10.1093/bioinformatics/btr629. Epub 2011 Nov 13. Bioinformatics. 2012. PMID: 22084253 Free PMC article.
-
SiNVICT: ultra-sensitive detection of single nucleotide variants and indels in circulating tumour DNA.Bioinformatics. 2017 Jan 1;33(1):26-34. doi: 10.1093/bioinformatics/btw536. Epub 2016 Aug 16. Bioinformatics. 2017. PMID: 27531099
-
Computational tools to detect signatures of mutational processes in DNA from tumours: A review and empirical comparison of performance.PLoS One. 2019 Sep 12;14(9):e0221235. doi: 10.1371/journal.pone.0221235. eCollection 2019. PLoS One. 2019. PMID: 31513583 Free PMC article. Review.
-
Base changes in tumour DNA have the power to reveal the causes and evolution of cancer.Oncogene. 2017 Jan 12;36(2):158-167. doi: 10.1038/onc.2016.192. Epub 2016 Jun 6. Oncogene. 2017. PMID: 27270430 Free PMC article. Review.
Cited by
-
De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly.Nat Genet. 2012 Jun 24;44(8):941-5. doi: 10.1038/ng.2329. Nat Genet. 2012. PMID: 22729223 Free PMC article.
-
MSIpred: a python package for tumor microsatellite instability classification from tumor mutation annotation data using a support vector machine.Sci Rep. 2018 Dec 3;8(1):17546. doi: 10.1038/s41598-018-35682-z. Sci Rep. 2018. PMID: 30510242 Free PMC article.
-
Identification of Somatic Mutations From Bulk and Single-Cell Sequencing Data.Front Aging. 2022 Jan 3;2:800380. doi: 10.3389/fragi.2021.800380. eCollection 2021. Front Aging. 2022. PMID: 35822012 Free PMC article. Review.
-
Postzygotic single-nucleotide mosaicisms in whole-genome sequences of clinically unremarkable individuals.Cell Res. 2014 Nov;24(11):1311-27. doi: 10.1038/cr.2014.131. Epub 2014 Oct 14. Cell Res. 2014. PMID: 25312340 Free PMC article.
-
A cancer cell-line titration series for evaluating somatic classification.BMC Res Notes. 2015 Dec 26;8:823. doi: 10.1186/s13104-015-1803-7. BMC Res Notes. 2015. PMID: 26708082 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

