Midbody assembly and its regulation during cytokinesis
- PMID: 22278743
- PMCID: PMC3302730
- DOI: 10.1091/mbc.E11-08-0721
Midbody assembly and its regulation during cytokinesis
Abstract
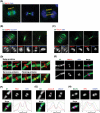
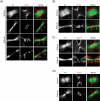
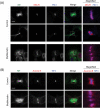
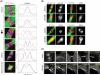
The midbody is a transient structure that connects two daughter cells at the end of cytokinesis, with the principal function being to localize the site of abscission, which physically separates two daughter cells. Despite its importance, understanding of midbody assembly and its regulation is still limited. Here we describe how the structural composition of the midbody changes during progression throughout cytokinesis and explore the functional implications of these changes. Deriving from midzones, midbodies are organized by a set of microtubule interacting proteins that colocalize to a zone of microtubule overlap in the center. We found that these proteins split into three subgroups that relocalize to different parts of the midbody: the bulge, the dark zone, and the flanking zone. We characterized these relocalizations and defined domain requirements for three key proteins: MKLP1, KIF4, and PRC1. Two cortical proteins-anillin and RhoA-localized to presumptive abscission sites in mature midbodies, where they may regulate the endosomal sorting complex required for transport machinery. Finally, we characterized the role of Plk1, a key regulator of cytokinesis, in midbody assembly. Our findings represent the most detailed description of midbody assembly and maturation to date and may help elucidate how abscission sites are positioned and regulated.
Figures


Similar articles
-
Kinesin-6 KIF20B is required for efficient cytokinetic furrowing and timely abscission in human cells.Mol Biol Cell. 2018 Jan 15;29(2):166-179. doi: 10.1091/mbc.E17-08-0495. Epub 2017 Nov 22. Mol Biol Cell. 2018. PMID: 29167382 Free PMC article.
-
The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules.J Cell Biol. 2013 Nov 11;203(3):505-20. doi: 10.1083/jcb.201306036. J Cell Biol. 2013. PMID: 24217623 Free PMC article.
-
Capping protein regulates actin dynamics during cytokinetic midbody maturation.Proc Natl Acad Sci U S A. 2018 Feb 27;115(9):2138-2143. doi: 10.1073/pnas.1722281115. Epub 2018 Feb 8. Proc Natl Acad Sci U S A. 2018. PMID: 29439200 Free PMC article.
-
The postmitotic midbody: Regulating polarity, stemness, and proliferation.J Cell Biol. 2019 Dec 2;218(12):3903-3911. doi: 10.1083/jcb.201906148. Epub 2019 Nov 5. J Cell Biol. 2019. PMID: 31690620 Free PMC article. Review.
-
Endocytic transport and cytokinesis: from regulation of the cytoskeleton to midbody inheritance.Trends Cell Biol. 2013 Jul;23(7):319-27. doi: 10.1016/j.tcb.2013.02.003. Epub 2013 Mar 20. Trends Cell Biol. 2013. PMID: 23522622 Free PMC article. Review.
Cited by
-
Coiled-coil domain containing protein 124 is a novel centrosome and midbody protein that interacts with the Ras-guanine nucleotide exchange factor 1B and is involved in cytokinesis.PLoS One. 2013 Jul 19;8(7):e69289. doi: 10.1371/journal.pone.0069289. Print 2013. PLoS One. 2013. PMID: 23894443 Free PMC article.
-
Recessive nephrocerebellar syndrome on the Galloway-Mowat syndrome spectrum is caused by homozygous protein-truncating mutations of WDR73.Brain. 2015 Aug;138(Pt 8):2173-90. doi: 10.1093/brain/awv153. Epub 2015 Jun 11. Brain. 2015. PMID: 26070982 Free PMC article.
-
Midbody Proteins Display Distinct Dynamics during Cytokinesis.Cells. 2022 Oct 22;11(21):3337. doi: 10.3390/cells11213337. Cells. 2022. PMID: 36359734 Free PMC article.
-
Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis.J Cell Biol. 2012 Sep 3;198(5):847-63. doi: 10.1083/jcb.201110060. J Cell Biol. 2012. PMID: 22945934 Free PMC article.
-
RNAi-mediated depletion of the NSL complex subunits leads to abnormal chromosome segregation and defective centrosome duplication in Drosophila mitosis.PLoS Genet. 2019 Sep 17;15(9):e1008371. doi: 10.1371/journal.pgen.1008371. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31527906 Free PMC article.
References
-
- Austin JR, 2nd, Segui-Simarro JM, Staehelin LA. Quantitative analysis of changes in spatial distribution and plus-end geometry of microtubules involved in plant-cell cytokinesis. J Cell Sci. 2005;118:3895–3903. - PubMed
-
- Boman AL, Kuai J, Zhu X, Chen J, Kuriyama R, Kahn RA. Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil Cytoskeleton. 1999;44:119–132. - PubMed
-
- Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

