Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine
- PMID: 22267340
- PMCID: PMC3336785
- DOI: 10.1096/fj.10-177980
Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine
Abstract
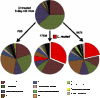
Beneficial microbes and probiotics show promise for the treatment of pediatric gastrointestinal diseases. However, basic mechanisms of probiosis are not well understood, and most investigations have been performed in germ-free or microbiome-depleted animals. We sought to functionally characterize probiotic-host interactions in the context of normal early development. Outbred CD1 neonatal mice were orally gavaged with one of two strains of human-derived Lactobacillus reuteri or an equal volume of vehicle. Transcriptome analysis was performed on enterocyte RNA isolated by laser-capture microdissection. Enterocyte migration and proliferation were assessed by labeling cells with 5-bromo-2'-deoxyuridine, and fecal microbial community composition was determined by 16S metagenomic sequencing. Probiotic ingestion altered gene expression in multiple canonical pathways involving cell motility. L. reuteri strain DSM 17938 dramatically increased enterocyte migration (3-fold), proliferation (34%), and crypt height (29%) compared to vehicle-treated mice, whereas strain ATCC PTA 6475 increased cell migration (2-fold) without affecting crypt proliferative activity. In addition, both probiotic strains increased the phylogenetic diversity and evenness between taxa of the fecal microbiome 24 h after a single probiotic gavage. These experiments identify two targets of probiosis in early development, the intestinal epithelium and the gut microbiome, and suggest novel mechanisms for probiotic strain-specific effects.
Figures



Similar articles
-
Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice.J Pediatr Gastroenterol Nutr. 2012 Sep;55(3):299-307. doi: 10.1097/MPG.0b013e31824d2548. J Pediatr Gastroenterol Nutr. 2012. PMID: 22343914 Free PMC article.
-
Human-derived probiotic Lactobacillus reuteri strains differentially reduce intestinal inflammation.Am J Physiol Gastrointest Liver Physiol. 2010 Nov;299(5):G1087-96. doi: 10.1152/ajpgi.00124.2010. Epub 2010 Aug 26. Am J Physiol Gastrointest Liver Physiol. 2010. PMID: 20798357 Free PMC article.
-
Administration of two probiotic strains during early childhood does not affect the endogenous gut microbiota composition despite probiotic proliferation.BMC Microbiol. 2017 Aug 17;17(1):175. doi: 10.1186/s12866-017-1090-7. BMC Microbiol. 2017. PMID: 28818050 Free PMC article.
-
Modulation of immunity and inflammatory gene expression in the gut, in inflammatory diseases of the gut and in the liver by probiotics.World J Gastroenterol. 2014 Nov 14;20(42):15632-49. doi: 10.3748/wjg.v20.i42.15632. World J Gastroenterol. 2014. PMID: 25400447 Free PMC article. Review.
-
Probiotic strains and mechanistic insights for the treatment of type 2 diabetes.Endocrine. 2017 Nov;58(2):207-227. doi: 10.1007/s12020-017-1433-z. Epub 2017 Oct 19. Endocrine. 2017. PMID: 29052181 Review.
Cited by
-
Increased in vitro migration of human umbilical cord mesenchymal stem cells toward acellular foreskin treated with bacterial derivatives of monophosphoryl lipid A or supernatant of Lactobacillus acidophilus.Hum Cell. 2020 Jan;33(1):10-22. doi: 10.1007/s13577-019-00308-7. Epub 2019 Dec 6. Hum Cell. 2020. PMID: 31811569
-
Interactions Between Commensal Bacteria and Enteric Neurons, via FPR1 Induction of ROS, Increase Gastrointestinal Motility in Mice.Gastroenterology. 2019 Jul;157(1):179-192.e2. doi: 10.1053/j.gastro.2019.03.045. Epub 2019 Mar 28. Gastroenterology. 2019. PMID: 30930024 Free PMC article.
-
Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species.EMBO J. 2013 Nov 27;32(23):3017-28. doi: 10.1038/emboj.2013.224. Epub 2013 Oct 18. EMBO J. 2013. PMID: 24141879 Free PMC article.
-
Conditioned medium from Bifidobacteria infantis protects against Cronobacter sakazakii-induced intestinal inflammation in newborn mice.Am J Physiol Gastrointest Liver Physiol. 2014 May 1;306(9):G779-87. doi: 10.1152/ajpgi.00183.2013. Epub 2014 Mar 13. Am J Physiol Gastrointest Liver Physiol. 2014. PMID: 24627567 Free PMC article.
-
Effect of Probiotics and Herbal Products on Intestinal Histomorphological and Immunological Development in Piglets.Vet Med Int. 2020 Apr 24;2020:3461768. doi: 10.1155/2020/3461768. eCollection 2020. Vet Med Int. 2020. PMID: 32373310 Free PMC article.
References
-
- Food and Agricultural Organization of the United Nations (FAO)/World Health Organization (WHO) (2001) Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria, FAO/WHO, Geneva, Switzerland
-
- Fuller R. (1989) Probiotics in man and animals. J. Appl. Bacteriol. 66, 365–378 - PubMed
-
- Thomas D. W., Greer F. R. (2010) Probiotics and prebiotics in pediatrics. Pediatrics 126, 1217–1231 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

