Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry
- PMID: 22253445
- PMCID: PMC3318727
- DOI: 10.1074/jbc.M111.312561
Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry
Abstract
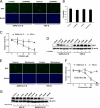
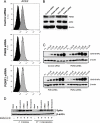
Phosphatidylinositol kinases (PI kinases) play an important role in the life cycle of several viruses after infection. Using gene knockdown technology, we demonstrate that phosphatidylinositol 4-kinase IIIβ (PI4KB) is required for cellular entry by pseudoviruses bearing the severe acute respiratory syndrome-coronavirus (SARS-CoV) spike protein and that the cell entry mediated by SARS-CoV spike protein is strongly inhibited by knockdown of PI4KB. Consistent with this observation, pharmacological inhibitors of PI4KB blocked entry of SARS pseudovirions. Further research suggested that PI4P plays an essential role in SARS-CoV spike-mediated entry, which is regulated by the PI4P lipid microenvironment. We further demonstrate that PI4KB does not affect virus entry at the SARS-CoV S-ACE2 binding interface or at the stage of virus internalization but rather at or before virus fusion. Taken together, these results indicate a new function for PI4KB and suggest a new drug target for preventing SARS-CoV infection.
Figures
Similar articles
-
SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway.Cell Res. 2008 Feb;18(2):290-301. doi: 10.1038/cr.2008.15. Cell Res. 2008. PMID: 18227861 Free PMC article.
-
Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry.Proc Natl Acad Sci U S A. 2004 Mar 23;101(12):4240-5. doi: 10.1073/pnas.0306446101. Epub 2004 Mar 9. Proc Natl Acad Sci U S A. 2004. PMID: 15010527 Free PMC article.
-
Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2.J Virol. 2010 Dec;84(24):12658-64. doi: 10.1128/JVI.01542-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926566 Free PMC article.
-
Coronavirus Spike Protein and Tropism Changes.Adv Virus Res. 2016;96:29-57. doi: 10.1016/bs.aivir.2016.08.004. Epub 2016 Sep 13. Adv Virus Res. 2016. PMID: 27712627 Free PMC article. Review.
-
Receptor recognition and cross-species infections of SARS coronavirus.Antiviral Res. 2013 Oct;100(1):246-54. doi: 10.1016/j.antiviral.2013.08.014. Epub 2013 Aug 29. Antiviral Res. 2013. PMID: 23994189 Free PMC article. Review.
Cited by
-
Reconstructed signaling and regulatory networks identify potential drugs for SARS-CoV-2 infection.bioRxiv [Preprint]. 2021 Dec 9:2020.06.01.127589. doi: 10.1101/2020.06.01.127589. bioRxiv. 2021. PMID: 33083801 Free PMC article. Preprint.
-
The function of apolipoproteins L (APOLs): relevance for kidney disease, neurotransmission disorders, cancer and viral infection.FEBS J. 2021 Jan;288(2):360-381. doi: 10.1111/febs.15444. Epub 2020 Jun 25. FEBS J. 2021. PMID: 32530132 Free PMC article. Review.
-
The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication.Biochem Pharmacol. 2012 Dec 1;84(11):1400-8. doi: 10.1016/j.bcp.2012.07.034. Epub 2012 Aug 8. Biochem Pharmacol. 2012. PMID: 22885339 Free PMC article. Review.
-
Lipidomics Issues on Human Positive ssRNA Virus Infection: An Update.Metabolites. 2020 Aug 31;10(9):356. doi: 10.3390/metabo10090356. Metabolites. 2020. PMID: 32878290 Free PMC article. Review.
-
Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks.Prog Lipid Res. 2013 Jul;52(3):294-304. doi: 10.1016/j.plipres.2013.04.002. Epub 2013 Apr 19. Prog Lipid Res. 2013. PMID: 23608234 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

