The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition
- PMID: 22244999
- PMCID: PMC3298764
- DOI: 10.1016/j.cub.2011.12.015
The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition
Abstract
Background: Induction of plant immune responses involves significant transcription reprogramming that prioritizes defense over growth-related cellular functions. Despite intensive forward genetic screens and genome-wide expression-profiling studies, a limited number of transcription factors have been found that regulate this transition.
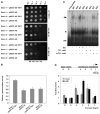
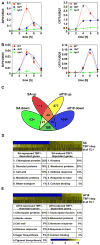
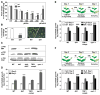
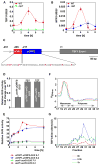
Results: Using the endoplasmic-reticulum-resident genes required for antimicrobial protein secretion as markers, we identified a heat-shock factor-like transcription factor that specifically binds to the TL1 (GAAGAAGAA) cis element required for the induction of these genes. Surprisingly, plants lacking this TL1-binding factor, TBF1, respond normally to heat stress but are compromised in immune responses induced by salicylic acid and by microbe-associated molecular pattern, elf18. Genome-wide expression profiling indicates that TBF1 plays a key role in the growth-to-defense transition. Moreover, the expression of TBF1 itself is tightly regulated at both the transcriptional and translational levels. Two upstream open reading frames encoding multiple aromatic amino acids were found 5' of the translation initiation codon of TBF1 and shown to affect its translation.
Conclusions: Through this unique regulatory mechanism, TBF1 can sense the metabolic changes upon pathogen invasion and trigger the specific transcriptional reprogramming through its target genes expression.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
uORF-mediated translation allows engineered plant disease resistance without fitness costs.Nature. 2017 May 25;545(7655):491-494. doi: 10.1038/nature22372. Epub 2017 May 17. Nature. 2017. PMID: 28514448 Free PMC article.
-
Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):126-32. doi: 10.1016/j.ejcb.2009.10.012. Epub 2009 Nov 27. Eur J Cell Biol. 2010. PMID: 19945192
-
Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis.Plant Physiol. 2012 May;159(1):52-5. doi: 10.1104/pp.111.191841. Epub 2012 Mar 16. Plant Physiol. 2012. PMID: 22427343 Free PMC article. No abstract available.
-
HSFA2 orchestrates transcriptional dynamics after heat stress in Arabidopsis thaliana.Transcription. 2016 Aug 7;7(4):111-4. doi: 10.1080/21541264.2016.1187550. Epub 2016 Jul 6. Transcription. 2016. PMID: 27383578 Free PMC article. Review.
-
Functions of heat shock transcription factors involved in response to photooxidative stresses in Arabidopsis.Biosci Biotechnol Biochem. 2016 Jul;80(7):1254-63. doi: 10.1080/09168451.2016.1176515. Epub 2016 Apr 20. Biosci Biotechnol Biochem. 2016. PMID: 27095030 Review.
Cited by
-
IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses.PLoS One. 2012;7(2):e31944. doi: 10.1371/journal.pone.0031944. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359644 Free PMC article.
-
Functionally coherent transcriptional responses of Jatropha curcas and Pseudomonas fragi for rhizosphere mediated degradation of pyrene.Sci Rep. 2024 Jan 10;14(1):1014. doi: 10.1038/s41598-024-51581-y. Sci Rep. 2024. PMID: 38200308 Free PMC article.
-
Long-distance communication and signal amplification in systemic acquired resistance.Front Plant Sci. 2013 Feb 22;4:30. doi: 10.3389/fpls.2013.00030. eCollection 2013. Front Plant Sci. 2013. PMID: 23440336 Free PMC article.
-
Metabolite Control of Translation by Conserved Peptide uORFs: The Ribosome as a Metabolite Multisensor.Plant Physiol. 2020 Jan;182(1):110-122. doi: 10.1104/pp.19.00940. Epub 2019 Aug 26. Plant Physiol. 2020. PMID: 31451550 Free PMC article.
-
Review: Emerging roles of the signaling network of the protein kinase GCN2 in the plant stress response.Plant Sci. 2022 Jul;320:111280. doi: 10.1016/j.plantsci.2022.111280. Epub 2022 Apr 1. Plant Sci. 2022. PMID: 35643606 Free PMC article. Review.
References
-
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. - PubMed
-
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

