Molecular determinants and dynamics of hepatitis C virus secretion
- PMID: 22241992
- PMCID: PMC3252379
- DOI: 10.1371/journal.ppat.1002466
Molecular determinants and dynamics of hepatitis C virus secretion
Abstract
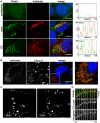
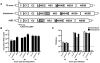
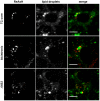
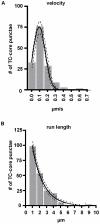
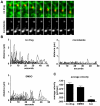
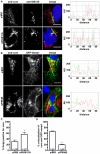
The current model of hepatitis C virus (HCV) production involves the assembly of virions on or near the surface of lipid droplets, envelopment at the ER in association with components of VLDL synthesis, and egress via the secretory pathway. However, the cellular requirements for and a mechanistic understanding of HCV secretion are incomplete at best. We combined an RNA interference (RNAi) analysis of host factors for infectious HCV secretion with the development of live cell imaging of HCV core trafficking to gain a detailed understanding of HCV egress. RNAi studies identified multiple components of the secretory pathway, including ER to Golgi trafficking, lipid and protein kinases that regulate budding from the trans-Golgi network (TGN), VAMP1 vesicles and adaptor proteins, and the recycling endosome. Our results support a model wherein HCV is infectious upon envelopment at the ER and exits the cell via the secretory pathway. We next constructed infectious HCV with a tetracysteine (TC) tag insertion in core (TC-core) to monitor the dynamics of HCV core trafficking in association with its cellular cofactors. In order to isolate core protein movements associated with infectious HCV secretion, only trafficking events that required the essential HCV assembly factor NS2 were quantified. TC-core traffics to the cell periphery along microtubules and this movement can be inhibited by nocodazole. Sub-populations of TC-core localize to the Golgi and co-traffic with components of the recycling endosome. Silencing of the recycling endosome component Rab11a results in the accumulation of HCV core at the Golgi. The majority of dynamic core traffics in association with apolipoprotein E (ApoE) and VAMP1 vesicles. This study identifies many new host cofactors of HCV egress, while presenting dynamic studies of HCV core trafficking in infected cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
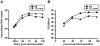



Similar articles
-
Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion.J Virol. 2016 Jul 27;90(16):7159-70. doi: 10.1128/JVI.00826-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27226379 Free PMC article.
-
Hepatitis C Virus Lipoviroparticles Assemble in the Endoplasmic Reticulum (ER) and Bud off from the ER to the Golgi Compartment in COPII Vesicles.J Virol. 2017 Jul 12;91(15):e00499-17. doi: 10.1128/JVI.00499-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28515296 Free PMC article.
-
Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion.J Biol Chem. 2012 Aug 10;287(33):27637-47. doi: 10.1074/jbc.M112.346569. Epub 2012 Jun 28. J Biol Chem. 2012. PMID: 22745132 Free PMC article.
-
Landscape of protein-protein interactions during hepatitis C virus assembly and release.Microbiol Spectr. 2024 Feb 6;12(2):e0256222. doi: 10.1128/spectrum.02562-22. Epub 2024 Jan 17. Microbiol Spectr. 2024. PMID: 38230952 Free PMC article. Review.
-
HCV egress - unconventional secretion of assembled viral particles.Trends Microbiol. 2022 Apr;30(4):364-378. doi: 10.1016/j.tim.2021.08.005. Epub 2021 Sep 2. Trends Microbiol. 2022. PMID: 34483048 Review.
Cited by
-
Nucleozin targets cytoplasmic trafficking of viral ribonucleoprotein-Rab11 complexes in influenza A virus infection.J Virol. 2013 Apr;87(8):4694-703. doi: 10.1128/JVI.03123-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408618 Free PMC article.
-
Understanding the hepatitis C virus life cycle paves the way for highly effective therapies.Nat Med. 2013 Jul;19(7):837-49. doi: 10.1038/nm.3248. Nat Med. 2013. PMID: 23836234 Free PMC article. Review.
-
Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion.J Virol. 2016 Jul 27;90(16):7159-70. doi: 10.1128/JVI.00826-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27226379 Free PMC article.
-
Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target.J Virol. 2012 Nov;86(21):11595-607. doi: 10.1128/JVI.01320-12. Epub 2012 Aug 15. J Virol. 2012. PMID: 22896614 Free PMC article.
-
Palmitoylation of Hepatitis C Virus NS2 Regulates Its Subcellular Localization and NS2-NS3 Autocleavage.J Virol. 2019 Dec 12;94(1):e00906-19. doi: 10.1128/JVI.00906-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597774 Free PMC article.
References
-
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

