A network flow approach to predict drug targets from microarray data, disease genes and interactome network - case study on prostate cancer
- PMID: 22239822
- PMCID: PMC3285036
- DOI: 10.1186/2043-9113-2-1
A network flow approach to predict drug targets from microarray data, disease genes and interactome network - case study on prostate cancer
Abstract
Background: Systematic approach for drug discovery is an emerging discipline in systems biology research area. It aims at integrating interaction data and experimental data to elucidate diseases and also raises new issues in drug discovery for cancer treatment. However, drug target discovery is still at a trial-and-error experimental stage and it is a challenging task to develop a prediction model that can systematically detect possible drug targets to deal with complex diseases.
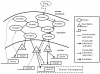
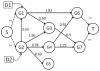
Methods: We integrate gene expression, disease genes and interaction networks to identify the effective drug targets which have a strong influence on disease genes using network flow approach. In the experiments, we adopt the microarray dataset containing 62 prostate cancer samples and 41 normal samples, 108 known prostate cancer genes and 322 approved drug targets treated in human extracted from DrugBank database to be candidate proteins as our test data. Using our method, we prioritize the candidate proteins and validate them to the known prostate cancer drug targets.
Results: We successfully identify potential drug targets which are strongly related to the well known drugs for prostate cancer treatment and also discover more potential drug targets which raise the attention to biologists at present. We denote that it is hard to discover drug targets based only on differential expression changes due to the fact that those genes used to be drug targets may not always have significant expression changes. Comparing to previous methods that depend on the network topology attributes, they turn out that the genes having potential as drug targets are weakly correlated to critical points in a network. In comparison with previous methods, our results have highest mean average precision and also rank the position of the truly drug targets higher. It thereby verifies the effectiveness of our method.
Conclusions: Our method does not know the real ideal routes in the disease network but it tries to find the feasible flow to give a strong influence to the disease genes through possible paths. We successfully formulate the identification of drug target prediction as a maximum flow problem on biological networks and discover potential drug targets in an accurate manner.
Figures





Similar articles
-
SCNrank: spectral clustering for network-based ranking to reveal potential drug targets and its application in pancreatic ductal adenocarcinoma.BMC Med Genomics. 2020 Apr 3;13(Suppl 5):50. doi: 10.1186/s12920-020-0681-6. BMC Med Genomics. 2020. PMID: 32241274 Free PMC article.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
Construction of a competing endogenous RNA network to identify drug targets against polycystic ovary syndrome.Hum Reprod. 2022 Nov 24;37(12):2856-2866. doi: 10.1093/humrep/deac218. Hum Reprod. 2022. PMID: 36223608
-
Exploring drug-target interaction networks of illicit drugs.BMC Genomics. 2013;14 Suppl 4(Suppl 4):S1. doi: 10.1186/1471-2164-14-S4-S1. Epub 2013 Oct 1. BMC Genomics. 2013. PMID: 24268016 Free PMC article. Review.
-
[Aiming for zero blindness].Nippon Ganka Gakkai Zasshi. 2015 Mar;119(3):168-93; discussion 194. Nippon Ganka Gakkai Zasshi. 2015. PMID: 25854109 Review. Japanese.
Cited by
-
IID 2021: towards context-specific protein interaction analyses by increased coverage, enhanced annotation and enrichment analysis.Nucleic Acids Res. 2022 Jan 7;50(D1):D640-D647. doi: 10.1093/nar/gkab1034. Nucleic Acids Res. 2022. PMID: 34755877 Free PMC article.
-
Drug2ways: Reasoning over causal paths in biological networks for drug discovery.PLoS Comput Biol. 2020 Dec 2;16(12):e1008464. doi: 10.1371/journal.pcbi.1008464. eCollection 2020 Dec. PLoS Comput Biol. 2020. PMID: 33264280 Free PMC article.
-
Network and matrix analysis of the respiratory disease interactome.BMC Syst Biol. 2014 Mar 22;8:34. doi: 10.1186/1752-0509-8-34. BMC Syst Biol. 2014. PMID: 24655443 Free PMC article.
-
GWAS and drug targets.BMC Genomics. 2014;15 Suppl 4(Suppl 4):S5. doi: 10.1186/1471-2164-15-S4-S5. Epub 2014 May 20. BMC Genomics. 2014. PMID: 25057111 Free PMC article.
-
Drug target prioritization by perturbed gene expression and network information.Sci Rep. 2015 Nov 30;5:17417. doi: 10.1038/srep17417. Sci Rep. 2015. PMID: 26615774 Free PMC article.
References
-
- Smith C. Hitting the target. Nature. 2003;422:341–347. - PubMed
-
- Lamb J, Crawford E, Peck D, Modell J, Blat I, Wrobel M, Lerner J, Brunet J, Subramanian A, Ross K, Reich M, Hieronymus H, Wei G, Armstrong S, Haggarty S, Clemons P, Wei R, Carr S, Lander E, Golub T. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. - DOI - PubMed
LinkOut - more resources
Full Text Sources

