Chlamydia trachomatis hijacks intra-Golgi COG complex-dependent vesicle trafficking pathway
- PMID: 22233276
- PMCID: PMC3330190
- DOI: 10.1111/j.1462-5822.2012.01747.x
Chlamydia trachomatis hijacks intra-Golgi COG complex-dependent vesicle trafficking pathway
Abstract
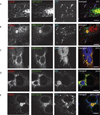
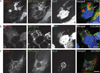
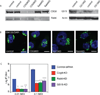
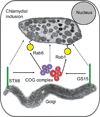
Chlamydia spp. are obligate intracellular bacteria that replicate inside the host cell in a bacterial modified unique compartment called the inclusion. As other intracellular pathogens, chlamydiae exploit host membrane trafficking pathways to prevent lysosomal fusion and to acquire energy and nutrients essential for their survival and replication. The Conserved Oligomeric Golgi (COG) complex is a ubiquitously expressed membrane-associated protein complex that functions in a retrograde intra-Golgi trafficking through associations with coiled-coil tethers, SNAREs, Rabs and COPI proteins. Several COG complex-interacting proteins, including Rab1, Rab6, Rab14 and Syntaxin 6 are implicated in chlamydial development. In this study, we analysed the recruitment of the COG complex and GS15-positive COG complex-dependent vesicles to Chlamydia trachomatis inclusion and their participation in chlamydial growth. Immunofluorescent analysis revealed that both GFP-tagged and endogenous COG complex subunits associated with inclusions in a serovar-independent manner by 8 h post infection and were maintained throughout the entire developmental cycle. Golgi v-SNARE GS15 was associated with inclusions 24 h post infection, but was absent on the mid-cycle (8 h) inclusions, indicating that this Golgi SNARE is directed to inclusions after COG complex recruitment. Silencing of COG8 and GS15 by siRNA significantly decreased infectious yield of chlamydiae. Further, membranous structures likely derived from lysed bacteria were observed inside inclusions by electron microscopy in cells depleted of COG8 or GS15. Our results showed that C. trachomatis hijacks the COG complex to redirect the population of Golgi-derived retrograde vesicles to inclusions. These vesicles likely deliver nutrients that are required for bacterial development and replication.
© 2012 Blackwell Publishing Ltd.
Conflict of interest statement
None declared.
Figures
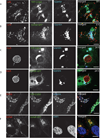
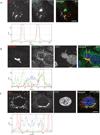
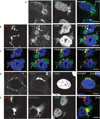
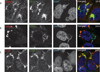

Similar articles
-
Chlamydia trachomatis intercepts Golgi-derived sphingolipids through a Rab14-mediated transport required for bacterial development and replication.PLoS One. 2010 Nov 22;5(11):e14084. doi: 10.1371/journal.pone.0014084. PLoS One. 2010. PMID: 21124879 Free PMC article.
-
The trans-Golgi SNARE syntaxin 10 is required for optimal development of Chlamydia trachomatis.Front Cell Infect Microbiol. 2015 Sep 25;5:68. doi: 10.3389/fcimb.2015.00068. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 26442221 Free PMC article.
-
Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner.Infect Immun. 2003 Oct;71(10):5855-70. doi: 10.1128/IAI.71.10.5855-5870.2003. Infect Immun. 2003. PMID: 14500507 Free PMC article.
-
Golgi inCOGnito: From vesicle tethering to human disease.Biochim Biophys Acta Gen Subj. 2020 Nov;1864(11):129694. doi: 10.1016/j.bbagen.2020.129694. Epub 2020 Jul 27. Biochim Biophys Acta Gen Subj. 2020. PMID: 32730773 Free PMC article. Review.
-
The Golgi puppet master: COG complex at center stage of membrane trafficking interactions.Histochem Cell Biol. 2013 Sep;140(3):271-83. doi: 10.1007/s00418-013-1117-6. Epub 2013 Jul 10. Histochem Cell Biol. 2013. PMID: 23839779 Free PMC article. Review.
Cited by
-
Isolation of Chlamydia trachomatis and membrane vesicles derived from host and bacteria.J Microbiol Methods. 2012 Nov;91(2):222-30. doi: 10.1016/j.mimet.2012.08.012. Epub 2012 Aug 30. J Microbiol Methods. 2012. PMID: 22960504 Free PMC article.
-
More than just sugars: Conserved oligomeric Golgi complex deficiency causes glycosylation-independent cellular defects.Traffic. 2018 Jun;19(6):463-480. doi: 10.1111/tra.12564. Epub 2018 Apr 24. Traffic. 2018. PMID: 29573151 Free PMC article.
-
Venous puncture wound hemostasis results in a vaulted thrombus structured by locally nucleated platelet aggregates.Commun Biol. 2021 Sep 16;4(1):1090. doi: 10.1038/s42003-021-02615-y. Commun Biol. 2021. PMID: 34531522 Free PMC article.
-
The Cytolytic Amphipathic β(2,2)-Amino Acid LTX-401 Induces DAMP Release in Melanoma Cells and Causes Complete Regression of B16 Melanoma.PLoS One. 2016 Feb 16;11(2):e0148980. doi: 10.1371/journal.pone.0148980. eCollection 2016. PLoS One. 2016. PMID: 26881822 Free PMC article.
-
Development and Initial Characterization of Cellular Models for COG Complex-Related CDG-II Diseases.Front Genet. 2021 Sep 17;12:733048. doi: 10.3389/fgene.2021.733048. eCollection 2021. Front Genet. 2021. PMID: 34603392 Free PMC article.
References
-
- Bruinsma P, Spelbrink RG, Nothwehr SF. Retrograde transport of the mannosyltransferase Och1p to the early Golgi requires a component of the COG transport complex. J Biol Chem. 2004;279:39814–39823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

